Weird `plot_ppc()` for numpyro model with custom mixture distribution
See original GitHub issueShort Description
I’m trying to estimate a finite mixture beta regression model using numpyro. The actual model I’m trying to fit is more complex, but I’ve reproduced my issue with a simpler model. y_obs is produced by two mixture components: one related to another variable x and another just centered at .50. This is roughly meant to represent how lots of human probability judgments look, where people are sometimes drawn to respond 50% when they are unsure.
Reproducible example and outputs
import numpyro
import numpyro.distributions as dist
from jax import numpy as jnp
from jax import random
from numpyro.infer import MCMC, NUTS, Predictive
import numpy as np
from numpy.random import default_rng
from jax.scipy.special import logit, expit
## simulate some data
n_obs = 2000
rg = default_rng(12345)
x = rg.normal(0,1,n_obs)
y_mean = expit(-.2 + .8*x)
y = rg.beta(y_mean*50, (1-y_mean)*50, n_obs)
p_drawn = .25
drawn = rg.binomial(1, p_drawn, n_obs)
dist50 = rg.beta(.5*300, .5*300, n_obs) # k = 300
y_obs = np.where(drawn==1, dist50, y)
The simulated data looks like this:
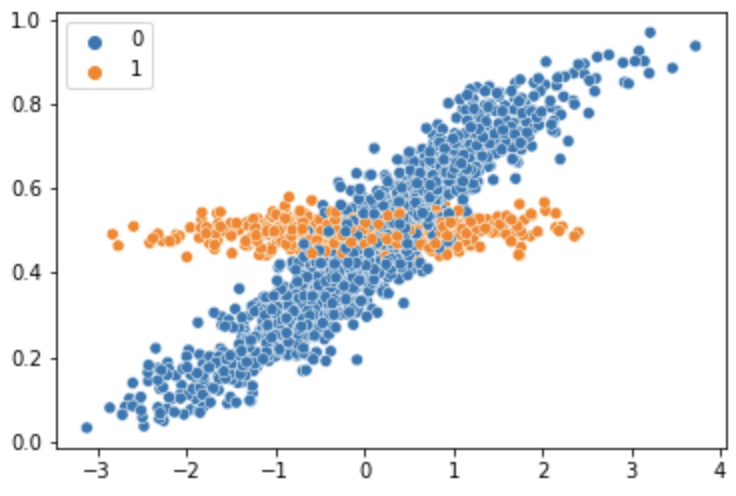
## define model
class MixtureBeta(dist.distribution.Distribution):
'''Mixture of Beta distribtions with marginalized latents
Takes a and b parameters for beta distributions along with a simplex mixing_probs parameter that defines the mixture probabilities.
'''
arg_constraints = {'concentration1': constraints.positive, 'concentration0': constraints.positive, 'mixing_probs': constraints.simplex}
reparametrized_params = ['concentration1', 'concentration0', 'mixing_probs']
support = constraints.unit_interval
def __init__(self, concentration1, concentration0, mixing_probs, validate_args=None):
self.concentration1, self.concentration0 = promote_shapes(concentration1, concentration0)
self.mixing_probs = jnp.reshape(mixing_probs, (1,-1))
batch_shape = lax.broadcast_shapes(jnp.shape(concentration1), jnp.shape(concentration0))
concentration1 = jnp.broadcast_to(concentration1, batch_shape)
concentration0 = jnp.broadcast_to(concentration0, batch_shape)
self._dirichlet = dist.Dirichlet(jnp.stack([concentration1, concentration0],
axis=-1))
super(MixtureBeta, self).__init__(batch_shape=batch_shape, validate_args=validate_args)
def sample(self, key, sample_shape=()):
assert is_prng_key(key)
samples = self._dirichlet.sample(key, sample_shape)[..., 0]
mixing_logits = jax.scipy.special.logit(self.mixing_probs)
ind = random.categorical(key, mixing_logits)
return samples[ind]
@validate_sample
def log_prob(self, value):
log_mixing_probs = jnp.log(self.mixing_probs.T)
dirichlet_probs = self._dirichlet.log_prob(jnp.stack([value, 1. - value], -1))
sum_probs = jnp.add(log_mixing_probs, dirichlet_probs)
return jax.nn.logsumexp(sum_probs, axis=0)
def mixmodel_beta_custom(x, y_obs=None):
alpha = numpyro.sample("alpha", dist.Normal(0,3))
beta = numpyro.sample("beta", dist.Normal(0,3))
mixing_probs = numpyro.sample("mixing", dist.Dirichlet(jnp.ones(2)))
k = numpyro.sample("k", dist.HalfCauchy(10))
mix_kval = numpyro.sample("mix_kval", dist.HalfCauchy(100))
mix_muval = numpyro.sample("mix_muval", dist.Beta(1,1))
y_true = expit(alpha + beta*x)
n_obs = y_true.shape[0]
with numpyro.plate("data", x.shape[0]):
yhat = jnp.stack([y_true, jnp.ones(n_obs)*mix_muval])
mix_k = jnp.stack([jnp.ones(n_obs)*k, jnp.ones(n_obs)*mix_kval])
numpyro.sample("yhat", MixtureBeta(yhat*mix_k, (1-yhat)*mix_k, mixing_probs), obs=y_obs)
## fit the model
kernel = NUTS(mixmodel_beta_custom)
mcmc_test = MCMC(kernel, 2000, 2000, num_chains=1)
mcmc_test.run(random.PRNGKey(0), x, y_obs)
## get posterior predictive
posterior_samples = mcmc_test.get_samples()
posterior_predictive = Predictive(mixmodel_beta_custom, posterior_samples)(
random.PRNGKey(1), x
)
prior = Predictive(mixmodel_beta_custom, num_samples=500)(
random.PRNGKey(10), x
)
az_data = az.from_numpyro(mcmc_test, prior=prior, posterior_predictive=posterior_predictive)
az.plot_ppc(data=az_data, var_names = "yhat", data_pairs={"yhat":"yhat"}, num_pp_samples=500)
This model recovers the correct parameters in the simulated data
mean std median 5.0% 95.0% n_eff r_hat
alpha -0.20 0.01 -0.20 -0.22 -0.19 2477.07 1.00
beta 0.79 0.01 0.79 0.78 0.81 2464.89 1.00
k 49.54 2.00 49.48 46.17 52.77 2153.06 1.00
mix_kval 317.33 29.35 317.21 268.25 365.02 1972.80 1.00
mix_muval 0.50 0.00 0.50 0.50 0.50 2539.01 1.00
mixing[0] 0.75 0.01 0.75 0.73 0.77 2612.54 1.00
mixing[1] 0.25 0.01 0.25 0.23 0.27 2612.54 1.00
but produces the funky plot below:
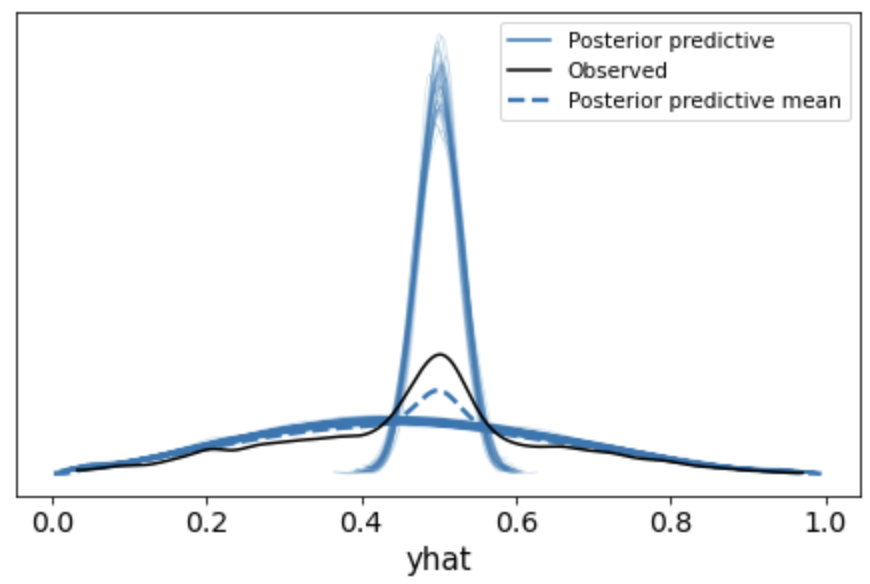
I’m wondering what i’m doing wrong? Unfortunately, I’m not sure if this is a numpyro or arviz issue, but thought I’d ask here first since the estimation seems to be working correctly but the plotting doesn’t.
Appreciate any help!
Issue Analytics
- State:
- Created 2 years ago
- Reactions:1
- Comments:8 (6 by maintainers)

 Top Related StackOverflow Question
Top Related StackOverflow Question
@derekpowell I just address the above issues and it seems to return the expected result. Could you double-check?
I think the takeaways here is to make sure that the implementation follows tensor shape senmatics: tensor/numpy arrays broadcast by aligning on the right. By the way, the above code can be generalized (using similar code) to more general patterns like
where we can replace
base_distwithBeta,Normal,… If you find that this Mixture class is useful, it would be great to open a FR or make a PR in numpyro. 😃Thanks! FYI, PyTorch has MixtureSameFamily that you can use as an API reference (the implementation there is a bit complicated to follow).