Could not obtain the same prediction results as represented in the paper.
See original GitHub issueHi CHENG, thanks for sharing the code.
I am running the code on the Combined Healthy Abdominal Organ Segmentation dataset. While after training, the result of the evaluation is quite different from that represented in the paper. I am wondering whether do I have some misoperation.
Here my steps are shown below:
At first, I downloaded datasets from https://chaos.grand-challenge.org/Download/. And I got datasets like this:
 Then as mentioned in the README.md file, I put the MR folder into
Then as mentioned in the README.md file, I put the MR folder into Self-supervised-Fewshot-Medical-Image-Segmentation/data/CHAOST2.
In the data pre-processing, first I run ./data/CHAOST2/dcm_img_to_nii.sh and ./data/CHAOST2/png_gth_to_nii.ipynp to convert dicom images to nifti files. After that, I got a folder named niis/T2SPIR, it consists of images in .nii.gz format.
 Then I run
Then I run ./data/CHAOST2/image_normalize.ipynb to do the pre-processing of the download images, and got normalized image folder chaos_MR_T2_normalized.
Next I run ./data/CHAOST2/class_slice_index_gen.ipynb to build class-slice indexing for setting up experiments.
Then to generate pseudo label, run ./data/pseudolabel_gen.ipynb, after all this done, the folder I got is shown below:
 Then I started to train the model by running
Then I started to train the model by running ./examples/train_ssl_abdominal_mri.sh and here my config file is shown below:
"""
Experiment configuration file
Extended from config file from original PANet Repository
"""
import os
import re
import glob
import itertools
import sacred
from sacred import Experiment
from sacred.observers import FileStorageObserver
from sacred.utils import apply_backspaces_and_linefeeds
from platform import node
from datetime import datetime
sacred.SETTINGS['CONFIG']['READ_ONLY_CONFIG'] = False
sacred.SETTINGS.CAPTURE_MODE = 'no'
ex = Experiment('mySSL')
ex.captured_out_filter = apply_backspaces_and_linefeeds
source_folders = ['.', './dataloaders', './models', './util']
sources_to_save = list(itertools.chain.from_iterable(
[glob.glob(f'{folder}/*.py') for folder in source_folders]))
for source_file in sources_to_save:
ex.add_source_file(source_file)
@ex.config
def cfg():
"""Default configurations"""
seed = 1234
gpu_id = 0
mode = 'train' # for now only allows 'train'
num_workers = 4 # 0 for debugging.
dataset = 'CHAOST2_Superpix' # i.e. abdominal MRI
use_coco_init = True # initialize backbone with MS_COCO initialization. Anyway coco does not contain medical images
### Training
n_steps = 100100
batch_size = 1
lr_milestones = [ (ii + 1) * 1000 for ii in range(n_steps // 1000 - 1)]
lr_step_gamma = 0.95
ignore_label = 255
print_interval = 100
save_snapshot_every = 25000
max_iters_per_load = 1000 # epoch size, interval for reloading the dataset
scan_per_load = -1 # numbers of 3d scans per load for saving memory. If -1, load the entire dataset to the memory
which_aug = 'sabs_aug' # standard data augmentation with intensity and geometric transforms
input_size = (256, 256)
min_fg_data='100' # when training with manual annotations, indicating number of foreground pixels in a single class single slice. This empirically stablizes the training process
label_sets = 0 # which group of labels taking as training (the rest are for testing)
exclude_cls_list = [2, 3] # testing classes to be excluded in training. Set to [] if testing under setting 1
usealign = True # see vanilla PANet
use_wce = True
### Validation
z_margin = 0
eval_fold = 0 # which fold for 5 fold cross validation
support_idx=[-1] # indicating which scan is used as support in testing.
val_wsize=2 # L_H, L_W in testing
n_sup_part = 3 # number of chuncks in testing
# Network
modelname = 'dlfcn_res101' # resnet 101 backbone from torchvision fcn-deeplab
clsname = None #
reload_model_path = None # path for reloading a trained model (overrides ms-coco initialization)
proto_grid_size = 8 # L_H, L_W = (32, 32) / 8 = (4, 4) in training
feature_hw = [32, 32] # feature map size, should couple this with backbone in future
# SSL
superpix_scale = 'MIDDLE' #MIDDLE/ LARGE
model = {
'align': usealign,
'use_coco_init': use_coco_init,
'which_model': modelname,
'cls_name': clsname,
'proto_grid_size' : proto_grid_size,
'feature_hw': feature_hw,
'reload_model_path': reload_model_path
}
task = {
'n_ways': 1,
'n_shots': 1,
'n_queries': 1,
'npart': n_sup_part
}
optim_type = 'sgd'
optim = {
'lr': 1e-3,
'momentum': 0.9,
'weight_decay': 0.0005,
}
exp_prefix = ''
exp_str = '_'.join(
[exp_prefix]
+ [dataset,]
+ [f'sets_{label_sets}_{task["n_shots"]}shot'])
path = {
'log_dir': './runs',
'SABS':{'data_dir': "/mnt/c/ubunturoot/Self-supervised-Fewshot-Medical-Image-Segmentation/data/SABS/sabs_CT_normalized"
},
'C0':{'data_dir': "feed your dataset path here"
},
'CHAOST2':{'data_dir': "/home/rc/Documents/Self-supervised-Fewshot-Medical-Image-Segmentation/data/CHAOST2/chaos_MR_T2_normalized/"
},
'SABS_Superpix':{'data_dir': "/mnt/c/ubunturoot/Self-supervised-Fewshot-Medical-Image-Segmentation/data/SABS/sabs_CT_normalized"},
'C0_Superpix':{'data_dir': "feed your dataset path here"},
'CHAOST2_Superpix':{'data_dir': "/home/rc/Documents/Self-supervised-Fewshot-Medical-Image-Segmentation/data/CHAOST2/chaos_MR_T2_normalized/"},
}
@ex.config_hook
def add_observer(config, command_name, logger):
"""A hook fucntion to add observer"""
exp_name = f'{ex.path}_{config["exp_str"]}'
observer = FileStorageObserver.create(os.path.join(config['path']['log_dir'], exp_name))
ex.observers.append(observer)
return config
After training, I have got some snapshot of the model:
 Then I run
Then I run ./examples/test_ssl_abdominal_mri.sh to test the model, and below is my result:
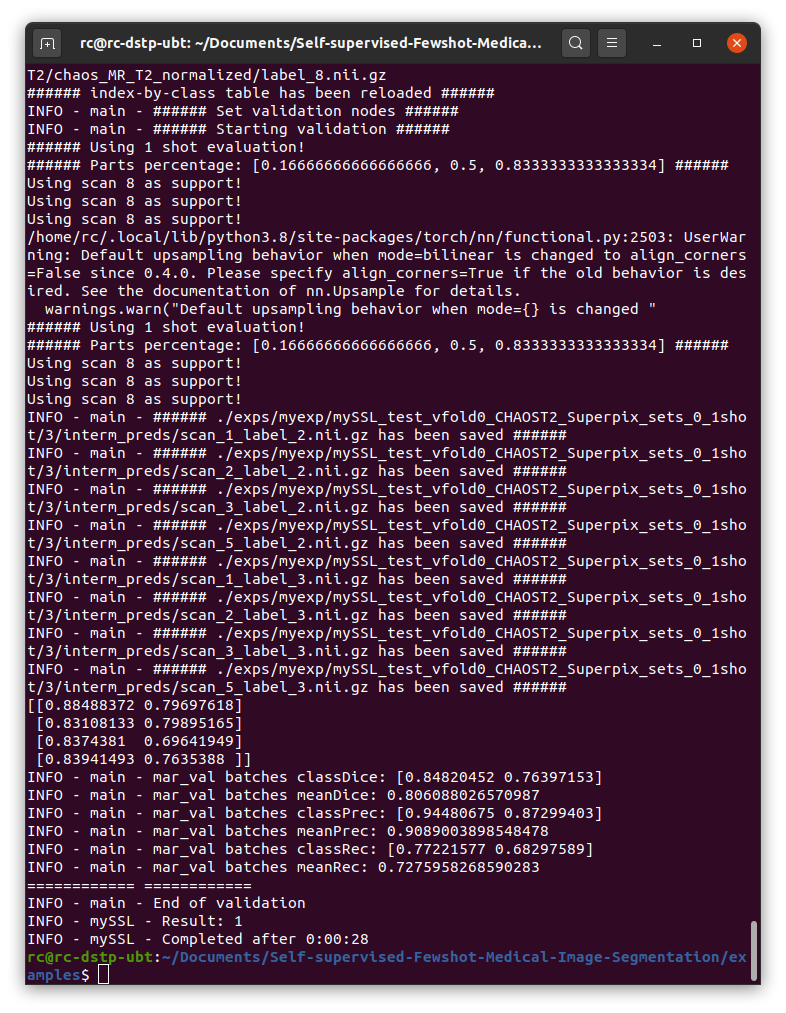 As is shown in the picture, the dice score is about 0.8, while in the paper, it shoule be around 80, it’s quite different.
So I wonder if there are some mistakes during my experiment? And how could I fix it and get ideal results?
I would deeply appreciate your reply.
As is shown in the picture, the dice score is about 0.8, while in the paper, it shoule be around 80, it’s quite different.
So I wonder if there are some mistakes during my experiment? And how could I fix it and get ideal results?
I would deeply appreciate your reply.
Issue Analytics
- State:
- Created 3 years ago
- Comments:33 (3 by maintainers)

 Top Related StackOverflow Question
Top Related StackOverflow Question
What has to be replaced in RELOAD_PATH ? Could anyone please clarify this ?
Hi, can you run CT test successfully? MRI test have no error, but when I test CT, the following problems arise: