[Bug] Posterior variance big compare to GPy for large N
See original GitHub issue🐛 Bug
I am trying to fit f(x) = sin(60 x⁴) with a gpytorch.kernels.ScaleKernel(gpytorch.kernels.RBFKernel()) kernel with many observations (N=1000). I have fixed the hyperparameters but the posterior variance seems to be much bigger than for GPy (see plots further down). We would expect it to approach zero everywhere for this many observations. Do you have a clue from where the instability could come?
To reproduce
** Code snippet to reproduce **
import math
import numpy as np
import torch
import gpytorch
from matplotlib import pyplot as plt
def gpytorch_model(noise=0.5, lengthscale=2.5, variance=2, n_iter=200):
bounds = np.array([[0,1]])
train_x = torch.linspace(bounds[0,0], bounds[0,1], 999).double()
train_y = np.sin(60 * train_x ** 4)
class ExactGPModel(gpytorch.models.ExactGP):
def __init__(self, train_x, train_y, likelihood):
super(ExactGPModel, self).__init__(train_x, train_y, likelihood)
self.mean_module = gpytorch.means.ZeroMean()
self.covar_module = gpytorch.kernels.ScaleKernel(
gpytorch.kernels.RBFKernel())
def forward(self, x):
mean_x = self.mean_module(x)
covar_x = self.covar_module(x)
return gpytorch.distributions.MultivariateNormal(mean_x, covar_x)
# initialize likelihood and model
likelihood = gpytorch.likelihoods.GaussianLikelihood()
model = ExactGPModel(train_x, train_y, likelihood)
model = model.double()
model.initialize(**{
'likelihood.noise': noise,
'covar_module.base_kernel.lengthscale': lengthscale,
'covar_module.outputscale': variance,
})
print("lengthscale: %.3f, variance: %.3f, noise: %.5f" % (model.covar_module.base_kernel.lengthscale.item(),
model.covar_module.outputscale.item(),
model.likelihood.noise.item()))
# Find optimal model hyperparameters
model.train()
likelihood.train()
# Use the adam optimizer
optimizer = torch.optim.Adam([
{'params': model.parameters()},
], lr=0.1)
# "Loss" for GPs - the marginal log likelihood
mll = gpytorch.mlls.ExactMarginalLogLikelihood(likelihood, model)
for i in range(n_iter):
# Zero gradients from previous iteration
optimizer.zero_grad()
# Output from model
output = model(train_x)
# Calc loss and backprop gradients
loss = -mll(output, train_y)
loss.backward()
optimizer.step()
# Prediction
# Get into evaluation (predictive posterior) mode
model.eval()
likelihood.eval()
# Test points are regularly spaced along [0,1]
# Make predictions by feeding model through likelihood
with torch.no_grad():
test_x = torch.linspace(bounds[0,0], bounds[0,1], 1000).double()
observed_pred = likelihood(model(test_x))
# Initialize plot
f, ax = plt.subplots(1, 1, figsize=(8, 3))
# Get upper and lower confidence bounds
var = observed_pred.variance.numpy()
mean = observed_pred.mean.numpy()
lower, upper = mean - 2 * np.sqrt(var), mean + 2 * np.sqrt(var)
# Plot training data as black stars
ax.plot(train_x.numpy(), train_y.numpy(), 'k*')
# Plot predictive means as blue line
ax.plot(test_x.numpy(), mean, 'b')
# Shade between the lower and upper confidence bounds
ax.fill_between(test_x.numpy(), lower, upper, alpha=0.5)
ax.legend(['Observed Data', 'Mean', 'Confidence'])
return model, {
'lengthscale': model.covar_module.base_kernel.lengthscale.item(),
'variance': model.covar_module.outputscale.item(),
'noise': model.likelihood.noise.item(),
}
model, params = gpytorch_model(noise=0.0001, lengthscale=0.015, variance=9, n_iter=0)
** Stack trace/error message **
Expected Behavior
We expect similar behaviour to GPy:

But got:

System information
GPyTorch Version: 0.3.2 PyTorch Version: 1.0.1.post2 Computer OS: MacOS 10.14
Additional context
Code to reproduce the GPy plot:
import math
import torch
import numpy as np
from matplotlib import pyplot as plt
import GPy
def gpy_model(noise=0.5, lengthscale=2.5, variance=2, optimize=True):
def f(x):
x = np.asanyarray(x)
y = where(x == 0, 1.0e-20, x)
return np.sin(y)/y
bounds = np.array([[0,1]])
train_x = torch.linspace(bounds[0,0], bounds[0,1], 999)
train_y = np.sin(60 * train_x ** 4)
kernel = GPy.kern.RBF(1, ARD=False)
model = GPy.models.GPRegression(train_x.numpy()[:,None], train_y.numpy()[:,None], kernel=kernel)
model.Gaussian_noise = noise
model.kern.lengthscale = lengthscale
model.kern.variance = variance
if optimize:
model.optimize()
test_x = np.linspace(bounds[0,0], bounds[0,1], 1000)
mean, var = model.predict(test_x[:, None])
mean = mean[:,0]
var = var[:,0]
# Initialize plot
f, ax = plt.subplots(1, 1, figsize=(8, 3))
# Get upper and lower confidence bounds
lower, upper = mean - 2 * np.sqrt(var), mean + 2 * np.sqrt(var)
# Plot training data as black stars
ax.plot(train_x.numpy(), train_y.numpy(), 'k*')
# Plot predictive means as blue line
ax.plot(test_x, mean, 'b')
# Shade between the lower and upper confidence bounds
ax.fill_between(test_x, lower, upper, alpha=0.5)
ax.legend(['Observed Data', 'Mean', 'Confidence'])
#print("l: {}, v: {}, noise: {}".format(kernel.lengthscale, kernel.variance, model.Gaussian_noise.variance))
return model, {
'lengthscale': kernel.lengthscale,
'noise': model.Gaussian_noise.variance,
'variance': kernel.variance
}
model, params = gpy_model(noise=0.0001, lengthscale=0.015, variance=9, optimize=False)
Issue Analytics
- State:
- Created 4 years ago
- Comments:7 (3 by maintainers)
 Top Results From Across the Web
Top Results From Across the Web
Posterior variance vs variance of the posterior mean
This question is about the frequentist properties of Bayesian methods. Suppose we have data y generated from a distribution with a single ...
Read more >Exact GP Regression with Multiple GPUs and Kernel Partitioning
In this notebook, we'll demonstrate training exact GPs on large datasets using two key ... The ability to distribute the kernel matrix across...
Read more >When Gaussian Process Meets Big Data - arXiv
It is worth noting that this review mainly focuses on scalable GPs for large-scale regression but not on all forms of GPs or...
Read more >Bayesian statistics and modelling - OSF
informative priors, owing to their larger variances. The mean hyperparameter can be seen as the ... posterior compared with an informative prior, depend-....
Read more >Bayesian Compression for Deep Learning - arXiv Vanity
While deep neural networks have become extremely successful in in a wide range ... of the approximate posterior over z suffers from high...
Read more > Top Related Medium Post
Top Related Medium Post
No results found
 Top Related StackOverflow Question
Top Related StackOverflow Question
No results found
 Troubleshoot Live Code
Troubleshoot Live Code
Lightrun enables developers to add logs, metrics and snapshots to live code - no restarts or redeploys required.
Start Free Top Related Reddit Thread
Top Related Reddit Thread
No results found
 Top Related Hackernoon Post
Top Related Hackernoon Post
No results found
 Top Related Tweet
Top Related Tweet
No results found
 Top Related Dev.to Post
Top Related Dev.to Post
No results found
 Top Related Hashnode Post
Top Related Hashnode Post
No results found

A few things:
K_{XX}^{-1}K_{XX*}solves for the predictive variances. In your case, addinggpytorch.settings.fast_pred_var()to the prediction context results in:The reason this works better is because in the very low noise setting you have extremely small eigenvalues that would be hard to get sufficient numerical accuracy for even in double precision; fast pred var runs until a low rank approximation to
K_{XX}+\sigma^{2}Iis sufficiently accurate, which effectively zeros out these eigenvalues.To answer your question in #727, turning on fast predictive variances also generally solved the issue with using fp32 as well (see below).
To answer your question about speed, in general, CG won’t be faster than Cholesky except (a) on a GPU (where you would already see substantial speed ups over Cholesky at n=1000), or (b) in very large data regimes where Cholesky simply cannot run due to the additional O(n^2) memory cost to store L (see e.g. the multi GPU / kernel checkpointing example notebook).
Of all the knobs available, for extremely numerically challenging problems, increasing the preconditioner size is indeed most likely to be successful. The convergence rate of CG improves exponentially for 1D RBF problems with the size of the preconditioner (Theorem 1 of the NeurIPS paper)
fp32 Code
Produces: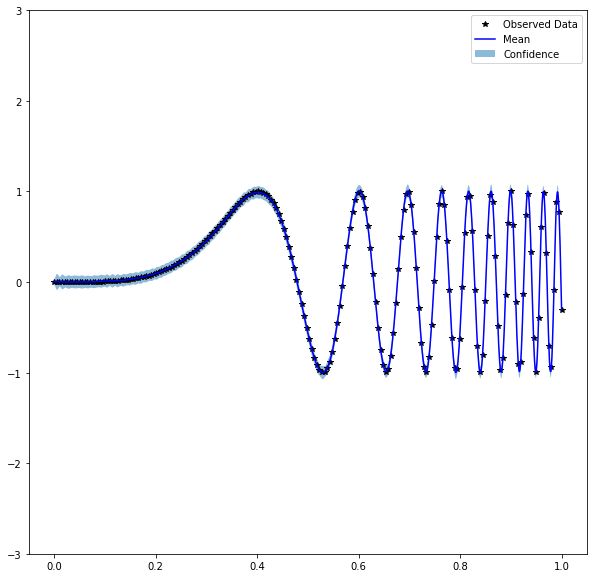
Yeah, so it’s a good question. I would conjecture that for something like the pivoted Cholesky decomposition or Lanczos, we could indeed prove exponential convergence in
||K - L_kL_k'||, where L_k is the decomposition (either from pivoted Cholesky, or by computingQT^{1/2}from Lanczos).In our paper, we observe at least empirically that, for example, CG can sometimes provide better solve residuals than Cholesky in fp32 (see Figure 2). This occurs for the same “regularizing” effect.
I think it is well known that this can occasionally happen in the scientific computing literature. I’ve asked @dme65 – we’ll see if we can dig up a reference for this phenomenon.