map_partitions or map_blocks with large objects eats up scheduler memory
See original GitHub issueOpening this report from investigation in https://github.com/dask/dask-ml/issues/842 and https://github.com/dask/dask-ml/pull/843
When passing some large objects as arguments to map_blocks / map_partitions, the scheduler memory can quickly be overwhelmed. Large objects should be wrapped in delayed to add them to graph and avoid this issue (see here) , but the way the object sizes are determined miss some large objects. In the cases I’ve encountered, it does not correctly compute the size of scikit-learn estimators.
This is because sys.getsizeof does not properly traverse object references to compute the “real” size of an object. This is a notoriously difficult thing to do in Python, so not sure what the best course of action here should be.
Minimal Complete Verifiable Example:
Running on machine with 2 cores and 16 GB of RAM
from dask_ml.datasets import make_classification
from sklearn.ensemble import RandomForestClassifier
import numpy as np
import pickle
import sys
import dask
from dask.distributed import Client
client = Client()
X, y = make_classification(
n_samples=50000,
chunks=1000,
random_state=42,
)
rf = RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)
_ = rf.fit(X, y)
def dask_predict(part, model):
return model.predict(part)
preds = X.map_blocks(
dask_predict,
model=rf,
dtype="int",
drop_axis=1,
)
preds.compute()
Scheduler memory ballons up to this:

And ends up with this error:
KilledWorker: ("('normal-dask_predict-4b858b85224825aeb2d45678c4c91d39', 27)", <WorkerState 'tcp://127.0.0.1:33369', name: 0, memory: 0, processing: 50>)
If we explictly delayed the rf object,
rf_delayed = dask.delayed(rf)
preds = X.map_blocks(
dask_predict,
model=rf_delayed,
dtype="int",
drop_axis=1,
meta=np.array([1]),
)
preds.compute()
the memory use looks like this:
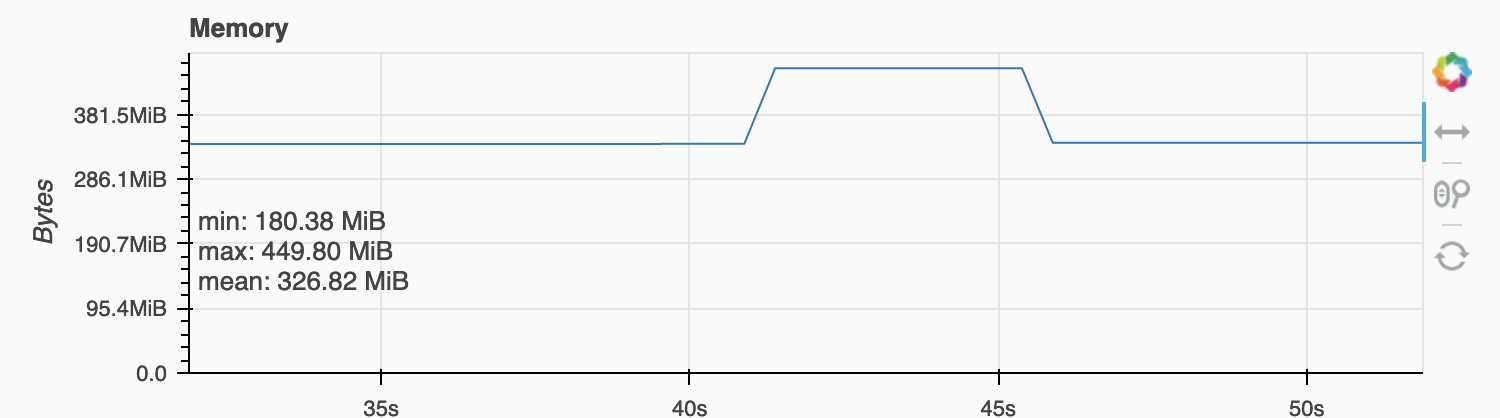
Environment:
- Dask version: 2021.5.1
- Python version: 3.7
- Operating System: ubuntu
- Install method (conda, pip, source): conda
Issue Analytics
- State:
- Created 2 years ago
- Comments:5 (4 by maintainers)

 Top Related StackOverflow Question
Top Related StackOverflow Question
Thanks for the clear writeup @rikturr! Indeed it looks like we’re underestimating the size of the
RandomForestClassifiermodel in your example:One way to improve the situation is to register a custom
sizeofimplementation indask/sizeof.pyfor scikit-learn estimators which more accurately captures the memory footprint of an estimator.One thing that comes to mind is to include information from
estimator.get_params()though there may be other attributes which are stored on the class, but not captured by
get_params.cc @thomasjpfan as you might find this interesting
I prefer Option 2b. It seems like a good idea to do as well as we can with the
sizeofestimation.