set_index on datetime column with microsecond precision removes one row from dataframe
See original GitHub issueWhat happened:
I created a pandas dataframe with shape (4, 2). One of the column is a datetime with microsecond precision.
If I create a dask dataframe from this pandas dataframe and call set_index on the datetime column, the resulting dataframe now has only 3 rows:
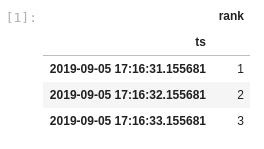
What you expected to happen:
If I comment out the line where we convert the index column to a datetime, then I get a dataframe with 4 rows:
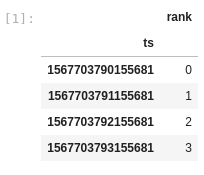
I expected the original code snippet to produce the same output (i.e. 4 row dataframe), but with the index as a datetime instead of a number of microseconds since 1970-01-01.
As a workaround, if I convert the column to datetime after setting the index, then I get what I really want:
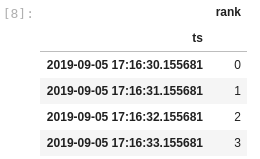
Minimal Complete Verifiable Example:
import dask.dataframe as dd
import pandas as pd
df = pd.DataFrame(
[
[1567703791155681, 1],
[1567703792155681, 2],
[1567703790155681, 0],
[1567703793155681, 3],
],
columns=["ts", "rank"]
)
df.ts = pd.to_datetime(df.ts, unit='us') # comment this line to get a df with 4 lines
ddf = dd.from_pandas(df, npartitions=2)
ddf = ddf.set_index("ts")
ddf.compute()
Anything else we need to know?:
I suspect the problem comes from the fact that the divisions of the dask dataframe after set_index are as follows:
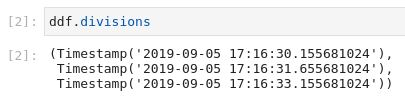
Notice that the divisions are precise to the nanosecond and that the first division is bigger than the timestamp of the row we’ve lost by 24 nanoseconds… Looks like numerical imprecision microseconds-to-datetime conversion maybe?
Environment:
- Dask version: 2.30.0
- Python version: 3.8.6
- Operating System: Linux (FROM python:3.8.6-buster in Dockerfile)
- Install method (conda, pip, source): pip
n.b. I ran the above snippet in a jupyter notebook.
Issue Analytics
- State:
- Created 3 years ago
- Comments:5 (3 by maintainers)

 Top Related StackOverflow Question
Top Related StackOverflow Question
Yeah - I wouldn’t be surprised if there were knock-on effects that I am not thinking of. I don’t feel confident that correcting the first division will cover all cases, but my intuition tells me that error in the last division isn’t likely to cause a problem. Note that any element falling beyond the threshold of the last division will be reassigned to the last partition anyway.
Alright, hopefully fixing the first division fixes the fact that the overall index is not sorted in the end… You certainly know better than I do! 😉
However, I am pretty sure (but not 100% certain) that I have also lost data (on my real 399 million rows dataframe) based on numerical imprecision of the last division… same problem with last and first divisions?
Oh, and thanks for such a fast analysis and answer to my issue… greatly appreciated!