QM9EdgeDataset labels are wrong
See original GitHub issue🐛 Bug
With QM9EdgeDataset, it seems that the prediction labels are broken. This may be because of the preprocessing, or because of a bad source for QM9.
To Reproduce
from dgl.data import QM9EdgeDataset as DGLQM9Edge
from dgl.data import QM9Dataset as DGLQM9
import matplotlib.pyplot as plt
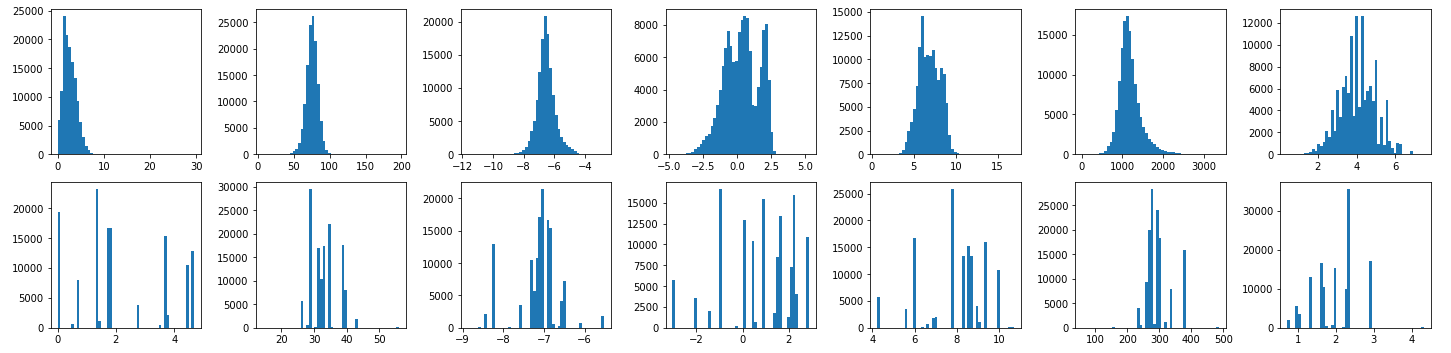
keys = ['mu', 'alpha', 'homo', 'lumo', 'gap', 'r2', 'zpve']
f, axs = plt.subplots(2, len(keys), figsize=(20, 5))
for i, task in enumerate(keys):
ds_dgl = DGLQM9Edge([task])
ds_dgl2 = DGLQM9([task])
targets_dgl = ds_dgl.targets[:,i]
targets_dgl2 =ds_dgl2.label[:,0]
axs[0][i].hist(targets_dgl2, bins=50)
axs[1][i].hist(targets_dgl, bins=50)
f.tight_layout()
plt.show()
The first row is the histograms of labels from QM9Dataset, and the second row is the ones from QM9EdgeDataset.
Expected behavior
Labels should be the same for all QM9 datasets.
Environment
- DGL Version (e.g., 1.0): commit 1f4c0b7
- Backend Library & Version (e.g., PyTorch 0.4.1, MXNet/Gluon 1.3): PyTorch 1.9.0
- OS (e.g., Linux): Linux
- How you installed DGL (
conda,pip, source): source - Build command you used (if compiling from source): cmake -DUSE_CUDA=ON -DUSE_FP16=ON … && make -j8
- Python version: 3.8.8
Additional context
In the docs it says that the preprocessing is done here https://gist.github.com/hengruizhang98/a2da30213b2356fff18b25385c9d3cd2 so there must be something wrong there.
Issue Analytics
- State:
- Created 2 years ago
- Comments:8 (8 by maintainers)
 Top Results From Across the Web
Top Results From Across the Web
QM9Dataset — DGL 0.8.2post1 documentation - DGL Docs
This dataset consists of 130,831 molecules with 12 regression targets. Nodes correspond to atoms and edges correspond to close atom pairs. This dataset...
Read more >Source code for schnetpack.datasets.qm9 - Read the Docs
The QM9 database contains small organic molecules with up to nine non-hydrogen ... functions to download QM9 from figshare and load the data...
Read more >QM7 dataset - Quantum-Machine.org
The energy and force labels for each geometry are included in the comment line ... Identifiers used in this data set agree with...
Read more >Adaptive Pseudo-labeling for Quantum Calculations
The challenge in pseudo-labeling is to prevent the bad pseudo-labels from biasing ... on the QM9 dataset with labels produced by density function...
Read more >16. Predicting DFT Energies with GNNs
QM9 is a dataset of 134,000 molecules consisting of 9 heavy atoms drawn from the ... There are multiple labels (see table below),...
Read more > Top Related Medium Post
Top Related Medium Post
No results found
 Top Related StackOverflow Question
Top Related StackOverflow Question
No results found
 Troubleshoot Live Code
Troubleshoot Live Code
Lightrun enables developers to add logs, metrics and snapshots to live code - no restarts or redeploys required.
Start Free Top Related Reddit Thread
Top Related Reddit Thread
No results found
 Top Related Hackernoon Post
Top Related Hackernoon Post
No results found
 Top Related Tweet
Top Related Tweet
No results found
 Top Related Dev.to Post
Top Related Dev.to Post
No results found
 Top Related Hashnode Post
Top Related Hashnode Post
No results found

Yes, It will take about 1 min to load graphs from QM9v2. While using QM9Edge is much faster as the graphs are constructed when called. You can choose the way you prefer.
I see that QM9V2 is directly loading DGL graphs with load_graphs, and QM9Edge is creating them on the fly. Maybe one is faster than the other.