SyntenyTrack not showing on browser
See original GitHub issueDescribe the bug
SyntenyTrack not showing after following the example in the combined guide to load a synteny track (https://jbrowse.org/jb2/docs/combined/)
To Reproduce
samtools 1.15.1
htslib 1.15.1
@jbrowse/cli/1.6.9 linux-x64 node-v14.18.3
minimap2 2.24-r1122
- Run the below commands:
jbrowse create local_jbrowse2 || (rm -rf local_jbrowse2 && jbrowse create local_jbrowse2)
samtools faidx grape.fasta
samtools faidx peach.fasta
minimap2 grape.fasta peach.fasta > peach_vs_grape.paf
jbrowse add-assembly grape.fasta --load copy --out local_jbrowse2 -n grape
jbrowse add-assembly peach.fasta --load copy --out local_jbrowse2 -n peach
jbrowse add-track peach_vs_grape.paf --assemblyNames peach,grape --out local_jbrowse2 --load copy
cd local_jbrowse2 && admin-server
- Go to the URL for jbrowse, add the query and target assemblies in the LinearSyntenyView:
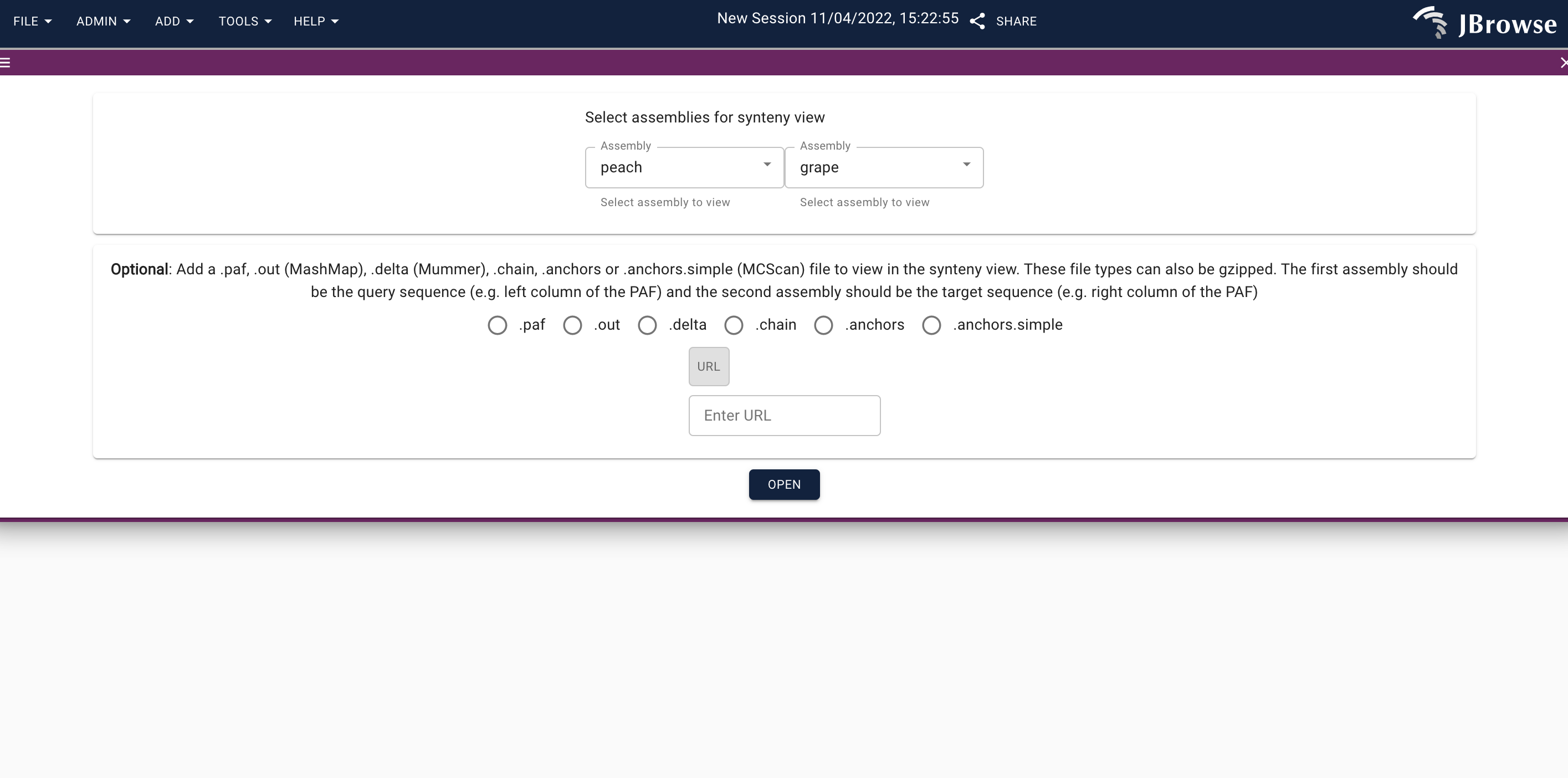
Please note I am leaving the .paf URL blank as the .paf file is already uploaded in the script above. This leads to the below:
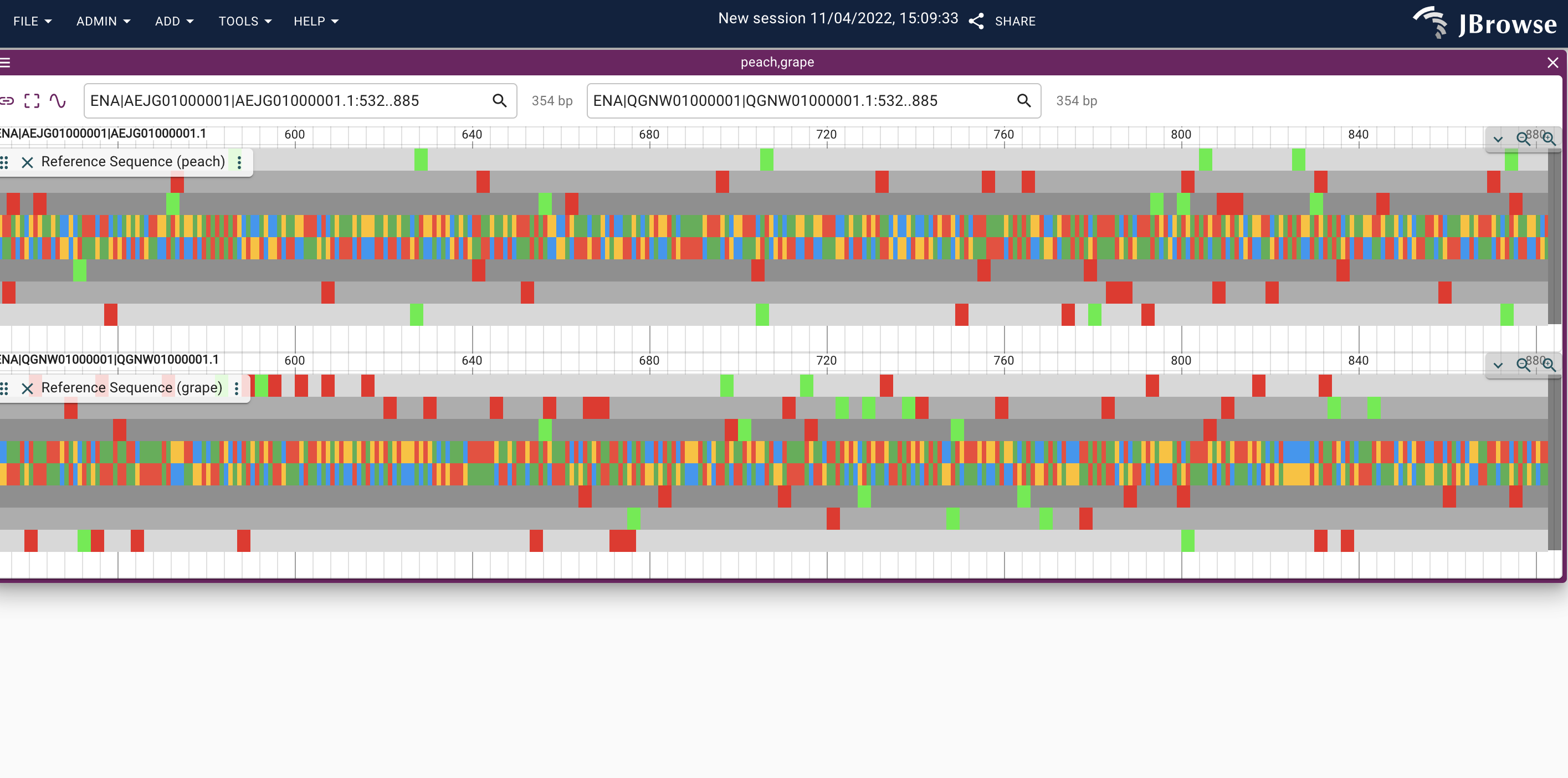
Expected behavior
I expect to see the red synteny track as below:
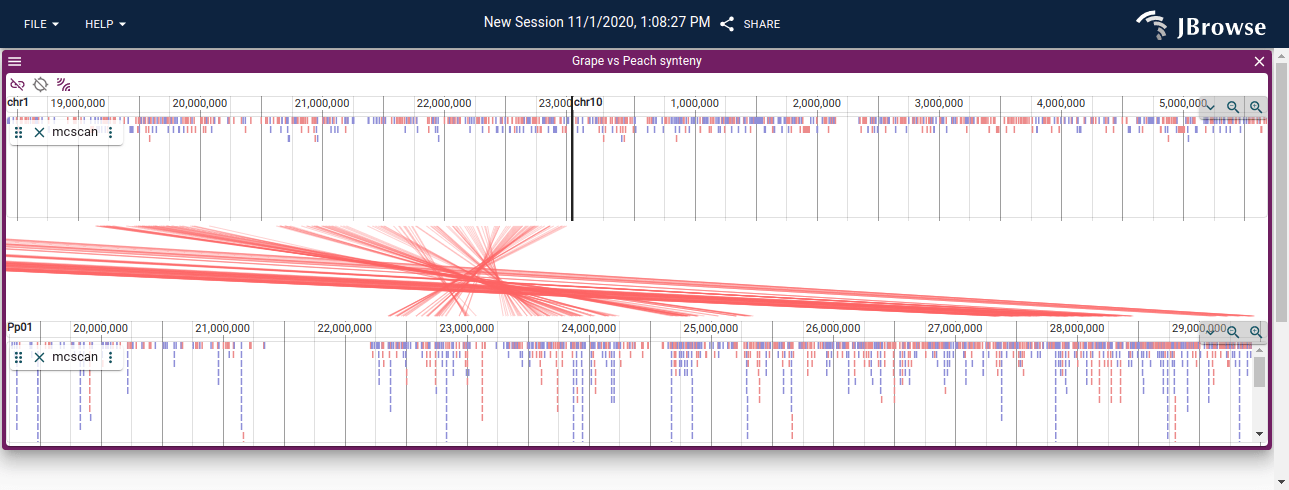
Version: 1.6.9, linux, installed with conda and running in docker, and accessed with Chrome
Issue Analytics
- State:
- Created a year ago
- Comments:5
 Top Results From Across the Web
Top Results From Across the Web
Synteny Tracks - Rat Genome Database
Synteny track popups contain additional information about the syntenic regions in both the “originating” species or assembly (i.e. the species or assembly whose ......
Read more >Genome Browser User's Guide
To control information overload, tracks need not be displayed in full. Tracks can be hidden, collapsed into a condensed or single-line display, ...
Read more >Synteny Browser Features
The syntenic block view browser displays the genome features in the reference and comparison species. This consists of five distinct elements (top to...
Read more >Using the Generic Synteny Browser (GBrowse_syn) - NCBI - NIH
Like GBrowse, GBrowse_syn can be configured to display any organism and is currently the synteny browser used for model organisms such as C....
Read more >JBrowse 2 guide
Adding a synteny track from a PAF file . ... Figure 2: JBrowse 2 screen showing no configuration found. Click on the sample...
Read more > Top Related Medium Post
Top Related Medium Post
No results found
 Top Related StackOverflow Question
Top Related StackOverflow Question
No results found
 Troubleshoot Live Code
Troubleshoot Live Code
Lightrun enables developers to add logs, metrics and snapshots to live code - no restarts or redeploys required.
Start Free Top Related Reddit Thread
Top Related Reddit Thread
No results found
 Top Related Hackernoon Post
Top Related Hackernoon Post
No results found
 Top Related Tweet
Top Related Tweet
No results found
 Top Related Dev.to Post
Top Related Dev.to Post
No results found
 Top Related Hashnode Post
Top Related Hashnode Post
No results found

Thanks! Please dont close the ticket yet - I am busy uploading my jbrowse folder to google drive to share - have some difficulties still.
Glad you got it to work! I definitely agree that adding some more comprehensive tutorial steps using the GUI would be very helpful. I wanted to do videos and maybe webpage tutorials here #2448 !