Simple Classification with MSE Loss
See original GitHub issueI’m getting hangups in the loss when doing classification using MSE loss. I’m attempting to follow the standard setup provided in the guides but the loss continues to get stuck at 0.25 regardless of the architecture of the network, which is making me suspicious that there is an error somewhere. The same setup works fine for a single dimensional regression task, so I believe the code is working.
I’ve attached a simple example trying to classify two dimensional data using MSE Loss
## IMPORTS ##
import math
import jax.numpy as np
from jax.experimental import stax
import jax
from jax import random
from jax import grad, jit, vmap
from jax.experimental import optimizers
## LOSS FUNCTION ##
def loss(params, apply_fn, inputs, targets):
preds = apply_fn(params, inputs)
out = np.mean((preds - targets)**2)
print(out)
return out
## INIT NETWORK ##
width = 100
init, apply = stax.serial(stax.Dense(width), stax.Relu,
stax.Dense(width), stax.Relu,
stax.Dense(width), stax.Relu,
stax.Dense(width), stax.Relu,
stax.Dense(1))
## GENERATE KEYS AND DATA ##
key = random.PRNGKey(10)
gen_key1, gen_key2, key = random.split(key, 3)
def twospirals(n_points, noise=.5, random_state=920):
"""
Returns the two spirals dataset.
"""
n = np.sqrt(random.uniform(key,(n_points,1))) * 600 * (2*np.pi)/360
d1x = -1.5*np.cos(n)*n + random.normal(gen_key1,(n_points,1)) * noise
d1y = 1.5*np.sin(n)*n + random.normal(gen_key2,(n_points,1)) * noise
return (np.vstack((np.hstack((d1x,d1y)),np.hstack((-d1x,-d1y)))),
np.hstack((np.zeros(n_points),np.ones(n_points))))
n_train = 500
train_x, train_y = twospirals(n_train, noise=0.5)
## DRAWING PARAMETERS ##
init_key = random.PRNGKey(42)
_, params = init(init_key, (-1, 2))
## RUNNING OPTIMIZATION ##
opt_init, opt_update, get_params = optimizers.adam(step_size=0.01)
def step(i, opt_state):
params = get_params(opt_state)
g = grad(loss)(params, apply, train_x, train_y)
return opt_update(i, g, opt_state)
# Optimize parameters in a loop
opt_state = opt_init(params)
for i in range(1000):
opt_state = step(i, opt_state)
And the classifier looks like:
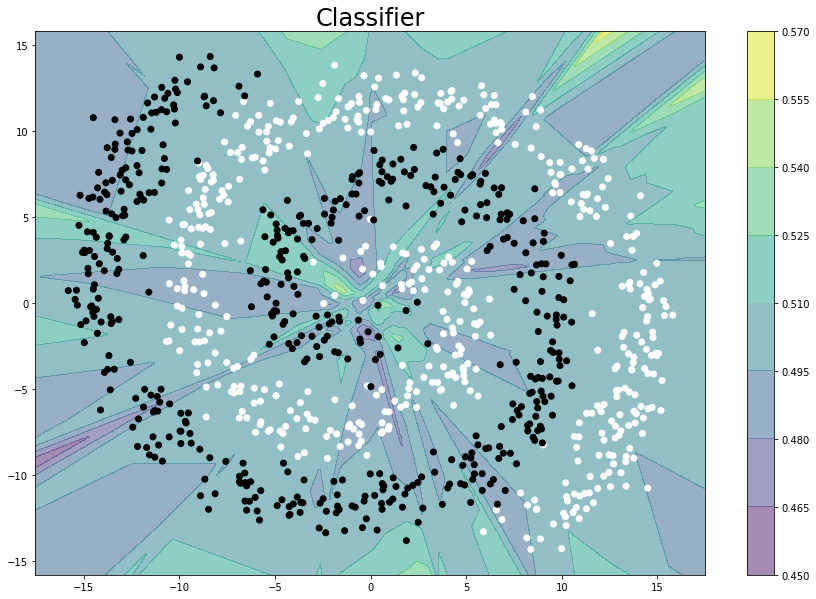
As mentioned above the fact that independent of model width/depth the loss does not go below 0.25 gives the sense that something is amiss. Any help would be appreciated - thanks!
Issue Analytics
- State:
- Created 3 years ago
- Reactions:1
- Comments:7 (4 by maintainers)
 Top Results From Across the Web
Top Results From Across the Web
Why Using Mean Squared Error(MSE) Cost Function for ...
There are two reasons why Mean Squared Error(MSE) is a bad choice for binary classification problems: First, using MSE means that we assume...
Read more >Can the mean squared error be used for classification?
Technically you can, but the MSE function is non-convex for binary classification.
Read more >Why not use mean squared error for classification problems?
According to the MSE, it's a perfect model, but, actually, it's not that good model, that's why we should not use MSE for...
Read more >Chapter 6 Logistic Regression 6.1 MSE / Convexity
In classification with MSE, the three requirements of a convex, ... With probabilities it is easy to recognize when a model has a...
Read more >Mean Squared Error vs Cross entropy loss function
Mean squared error (MSE) loss is used in regression tasks where we are trying to minimize an expected value of some function on...
Read more > Top Related Medium Post
Top Related Medium Post
No results found
 Top Related StackOverflow Question
Top Related StackOverflow Question
No results found
 Troubleshoot Live Code
Troubleshoot Live Code
Lightrun enables developers to add logs, metrics and snapshots to live code - no restarts or redeploys required.
Start Free Top Related Reddit Thread
Top Related Reddit Thread
No results found
 Top Related Hackernoon Post
Top Related Hackernoon Post
No results found
 Top Related Tweet
Top Related Tweet
No results found
 Top Related Dev.to Post
Top Related Dev.to Post
No results found
 Top Related Hashnode Post
Top Related Hashnode Post
No results found

I suspect that the predictions and the targets have different dimensions in the loss function. Let’s say N is
width. The output of theDense(1)layer is 2-dimensional, N x 1. Meanwhile, the targets that you supply are 1-dimensional, of length N. This causestargetsto be broadcast withinloss, but likely not in the way that you intend:preds - targetsis (N, N)-shaped.This behavior is dictated by NumPy, so the same issue would have come up in a pure NumPy implementation, without having JAX involved.
You could change the training set to produce (N, 1)-shaped targets. Alternatively, in
loss, changing the lineto
should work as well.
@hawkinsp’s suggestion above likely improved matters simply because it led to 2-dimensional targets.
Hello @froystig ,
Your comment above about preds dimension being (N,1) in loss function helped me a lot. I was stuck on it for long as my MSE for simple regression task was not reducing beyond particular value. With just addition of squeeze() solved problem immediately. I had no idea that broadcasting can create such problem. I was just doing .mean() on last value without checking dimensions.
Your one comment helped me solve my problem.
Really Appreciate it.
Regards, Sunny