Inference on images with shapes different from training
See original GitHub issueIssue description
This issue has for goal to document inference on images with dimensions different from the ones used during training.
2D UNet
The 2D model trained for spinal cord segmentation in the module sct_deepseg named seg_sc_t2star was used for testing.
- I ran the original model with the following command line. The resulting segmentation is presented in red (see image).
sct_deepseg -i data_example_spinegeneric/sub-cardiff03/anat/sub-cardiff03_T2star.nii.gz -o cropping.nii.gz -task seg_sc_t2star
- I locally modified the configuration file in
spinalcordtoolbox/data/deepseg_models/t2star_sc/t2star_sc.jsonand removed the cropping step that center cropped the image to a 160x160 pixel image. Each image passed through the model was now 307x307 pixels. I re-run the command listed in step 1. The resulting segmentation is presented in blue.
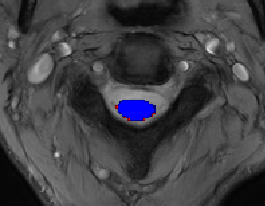
Observation: It was possible to run the model on images of the cropped image of size 160x160 and on the whole image of 307x307 pixels. Both models have similar outputs (but not exactly the same).
3D UNet
The task trained for spinal cord tumor segmentation composed of 2 cascaded model (1-SC detection and cropping and 2-Tumor segmentation) in the module sct_deepseg named seg_tumor-edema-cavity_t1-t2 was used for testing.
- I ran the original model with the following command line.
sct_deepseg -i 'sub-Astr144/anat/sub-Astr144_T2w.nii.gz' 'sub-Astr144/anat/sub-Astr144_T1w.nii.gz' -c t2 t1 -o cropping.nii.gz -task seg_tumor-edema-cavity_t1-t2
- I locally modified the configuration file in
spinalcordtoolbox/data/deepseg_models/model_seg_sctumor-edema-cavity_t2-t1_unet3d-multichannel/model_seg_sctumor-edema-cavity_t2-t1_unet3d-multichannel.jsonand modified the length and stride of patches to match the size of the resampled image (240x240x19). This way, only one big patch would be processed at once. I run the command following the command line to run only the tumor segmentation model (without the initial cropping).
sct_deepseg -i 'sub-Astr144/anat/sub-Astr144_T2w.nii.gz' 'sub-Astr144/anat/sub-Astr144_T1w.nii.gz' -c t2 t1 -o no_cropping.nii.gz -task model_seg_sctumor-edema-cavity_t2-t1_unet3d-multichannel
I encountered the following error:
File "/home/GRAMES.POLYMTL.CA/anlemj/ivadomed/ivadomed/models.py", line 1078, in forward
out = torch.cat([out, context_4], dim=1)
RuntimeError: Sizes of tensors must match except in dimension 4. Got 3 and 4
The error is caused by a mismatch in the tensor’s last dimension during the decoding part ([1, 128, 30, 30, 3] instead of [1, 128, 30, 30, 4] as we have 4 classes in this model ) of our modified 3D UNet. There is probably an operation that is incompatible with this manipulation in the way ivadomed’s 3D UNet is implemented.
@mariehbourget also tested this on microscopy images. Details to come.
Issue Analytics
- State:
- Created 2 years ago
- Reactions:4
- Comments:7 (7 by maintainers)

 Top Related StackOverflow Question
Top Related StackOverflow Question
Thank you @andreanne-lemay for opening the issue. Here are the results from my tests on microscopy models trained with 2D patches.
2D UNet trained with patches on SEM and TEM microscopy data
I tested the SEM and TEM axon-myelin segmentation models. The 2D model were trained with patches of 256x256 and 512x512 respectively, without any cropping.
I ran locally the ivadomed
--segmentcommand on the original model. The images are loaded as several patches and the prediction image is recontructed afterwards.I modified the configuration file in the model folder and removed the
length_2Dandstride_2Dparameters fromdefault_model. Each image are loaded as 1 slice at the original size of the image.Example images from the SEM model: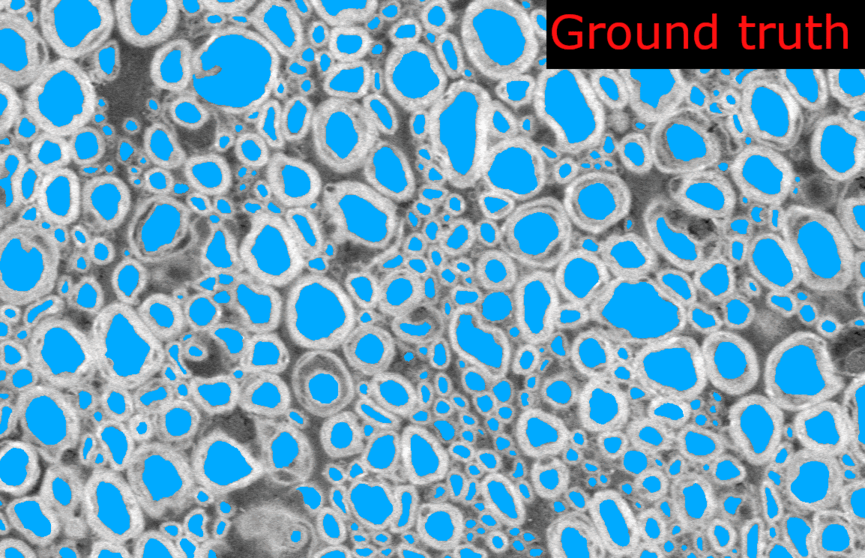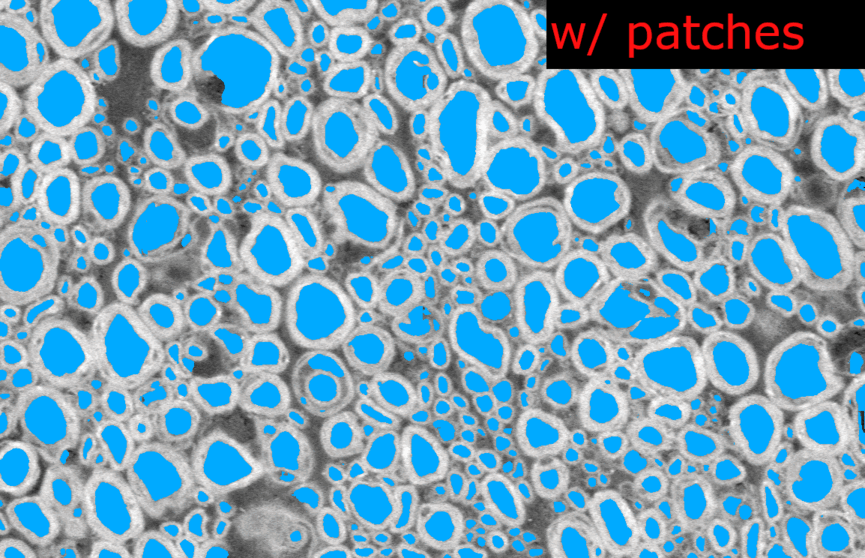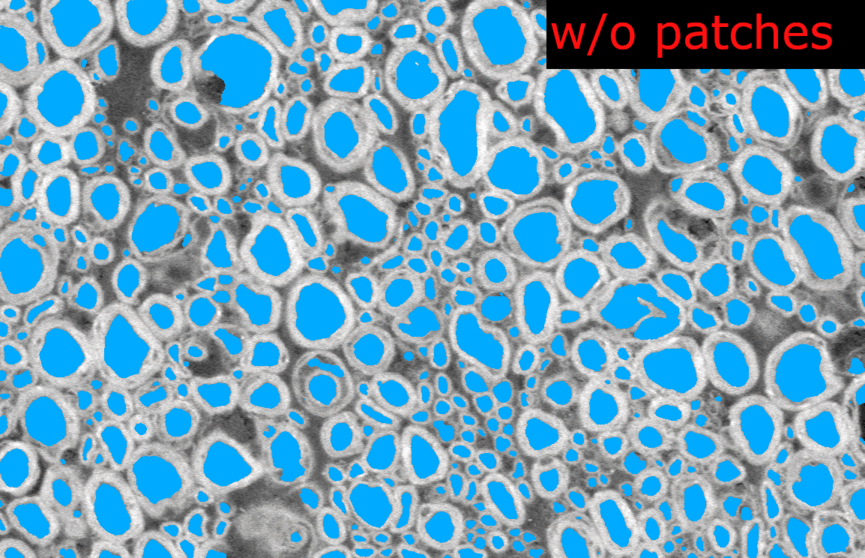
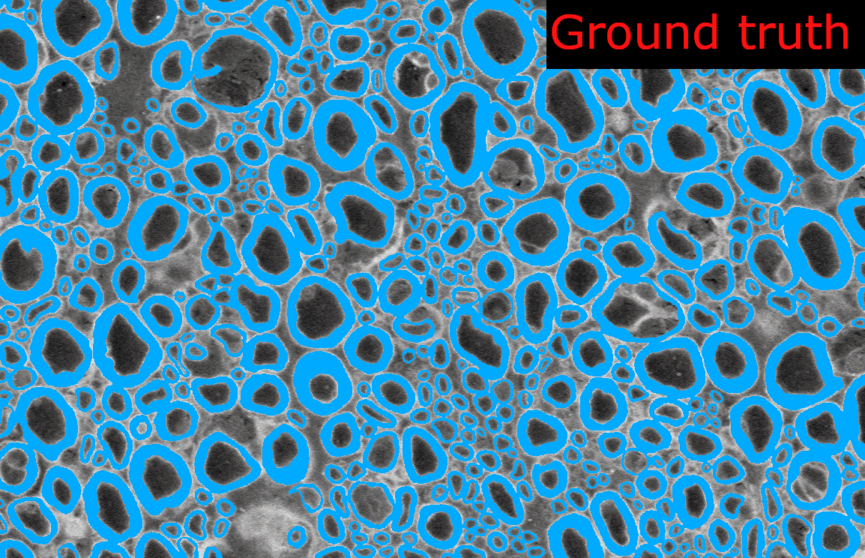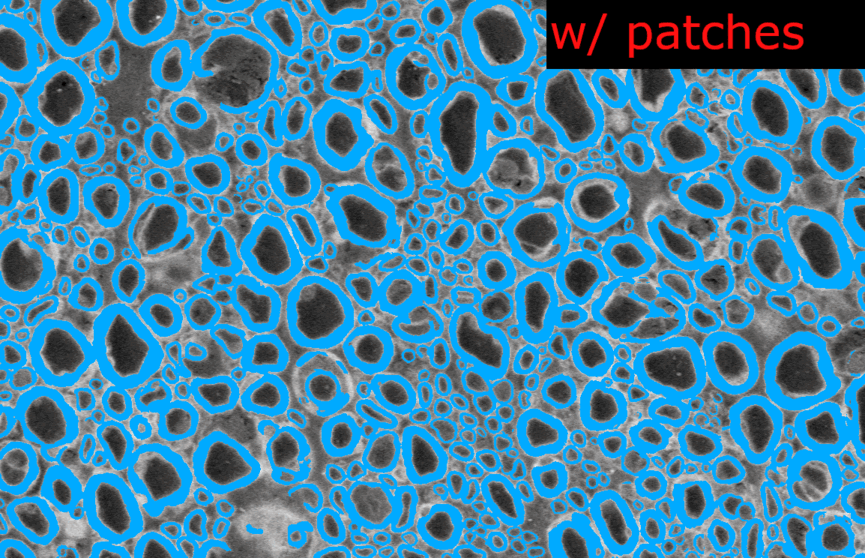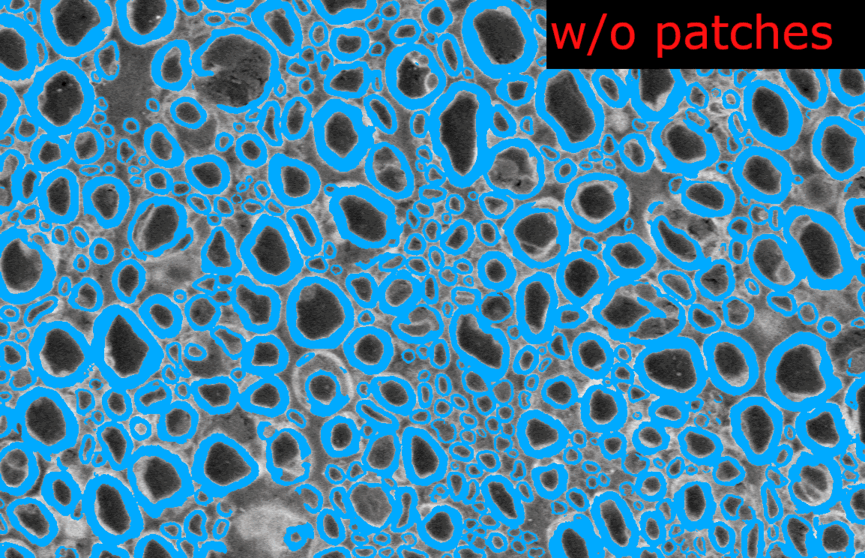
And the corresponding metrics: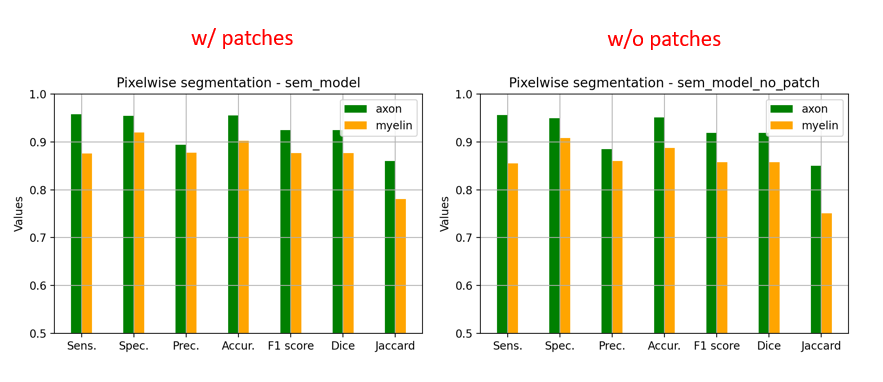
Observations:
2D UNet trained with patches on BF (bright-field optical) microscopy data
I also tested the BF axon-myelin segmentation models. The 2D model was trained with patches of 512x512, and I did the same procedure as above to segment with and without patches.
Observations:
I added some images and metrics above for the SEM model.
In the model without patches, the results seems slightly better visually because there is no artefact of “patch pattern” (especially for axons) but the Dice is slightly lower (especially for myelin). This is only on one test image for each model, I don’t have the bandwidth to test further this week. There is no clear “winner” but my intuition is that we would globally get better segmentation results without using patches at inference time.
As you mentioned, we would need to have the option to segment with or without the patches because of RAM on large microscopy images.