When representing interactions, is it possible to exclude interactions among amino acids?
See original GitHub issueWhen using docking-viewer example to represent interactions, is it possible to programmatically control Mol* to only show interactions between polymer and ligand, and do not show interactions among amino acids (e.g. interactions in the red box shown below)?
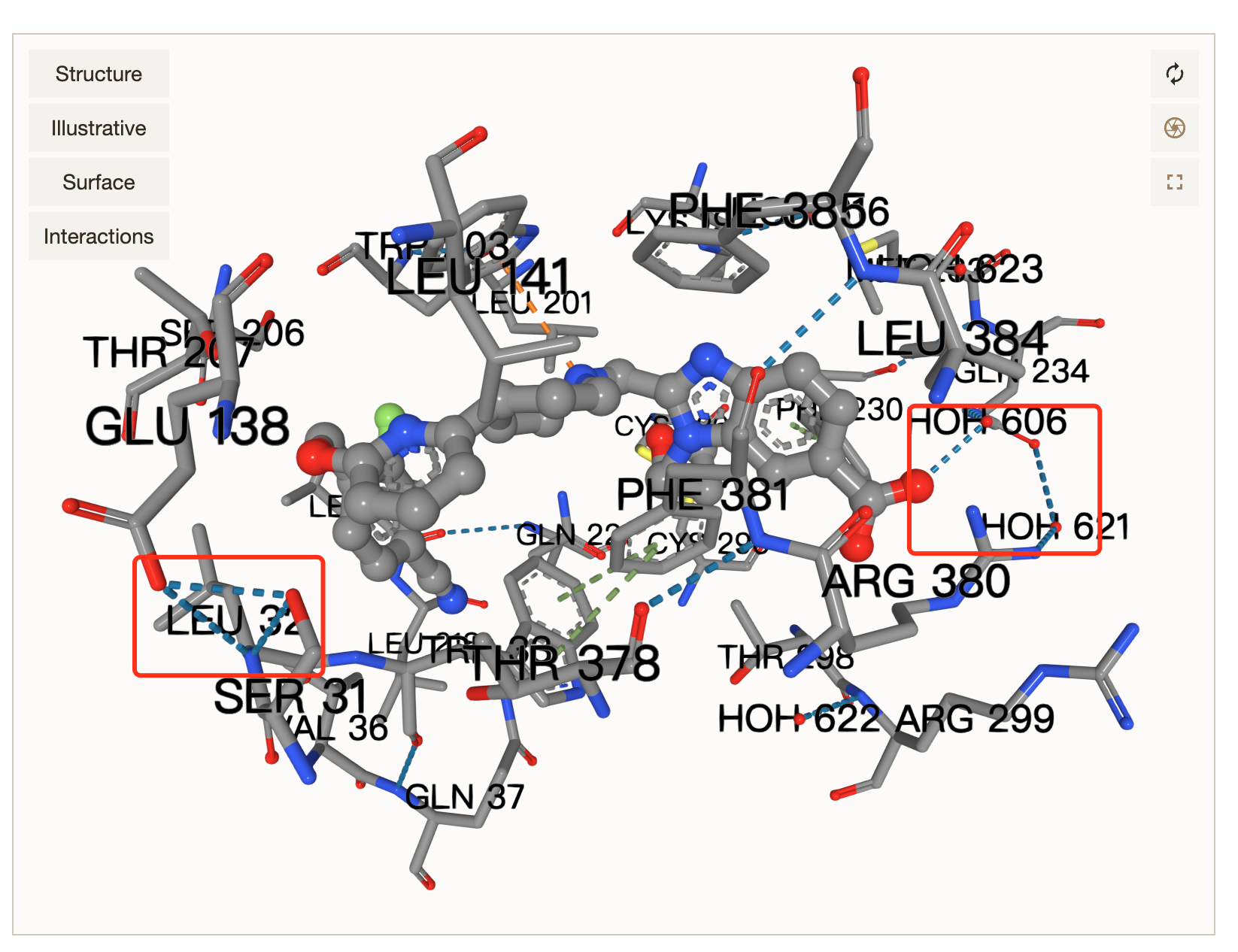
It seems that this function determines computing process of interactions, maybe I should override this file and add some extra filtering logics here? If it’s not too much trouble, can you give some code example to do so?
Thanks!
Issue Analytics
- State:
- Created a year ago
- Comments:10 (5 by maintainers)
 Top Results From Across the Web
Top Results From Across the Web
Amino acid–base interactions: a three-dimensional analysis of ...
They proposed a scheme whereby certain nucleotides could be recognised by particular amino acid side chains and reasoned that greater specificity was more ......
Read more >Orders of protein structure: primary, secondary, tertiary, and ...
Each chain has its own set of amino acids, assembled in a particular order. ... R group interactions that contribute to tertiary structure...
Read more >Cation-π interactions in structural biology - PNAS
Cation-π interactions in protein structures are identified and evaluated by using an energy-based criterion for selecting significant sidechain pairs.
Read more >Four Distances between Pairs of Amino Acids Provide ... - PLOS
The three-dimensional structures of proteins are stabilized by the interactions between amino acid residues. Here we report a method where ...
Read more >Coevolution-based inference of amino acid interactions ... - eLife
We created a library of all possible single and double mutations in the nine-residue α2 helix of five sequence-diverged PDZ homologs (PSD95 pdz3, ......
Read more > Top Related Medium Post
Top Related Medium Post
No results found
 Top Related StackOverflow Question
Top Related StackOverflow Question
No results found
 Troubleshoot Live Code
Troubleshoot Live Code
Lightrun enables developers to add logs, metrics and snapshots to live code - no restarts or redeploys required.
Start Free Top Related Reddit Thread
Top Related Reddit Thread
No results found
 Top Related Hackernoon Post
Top Related Hackernoon Post
No results found
 Top Related Tweet
Top Related Tweet
No results found
 Top Related Dev.to Post
Top Related Dev.to Post
No results found
 Top Related Hashnode Post
Top Related Hashnode Post
No results found

Well, seems like it got the job done for you and the solution looks sounds. We might discuss with @arose later this if we include this to the main library.
Hello everyone, the topic of current issue is very helpful for me. But I have a question about docking-viewer, how to toggle the docking-viewer option? I did’t see the toggle button on the UI.
Thanks a lot!
@arose @xuzuodong