Display cortical parcellation values: transparent medial wall and threshold both negative and positive values
See original GitHub issueHi,
I used on OSX pysurfer=0.11.dev0 with python=3.6.10 and tried to display on Desikan parcellation values, containing both negative and positives values, using the following code:
coef_lh_parcel = np.array([ 0.31447462, -0.08999642, 0.20214564, -0.05422508, 0. ,
-0.27036938, 0.27037478, -0.11377747, -0.07266576, -0.08349062,
0.01729273, -0.14993414, 0.04560162, -0.01732918, 0.1315399 ,
0.01141919, -0.02154149, -0.12041933, 0.26752043, 0.05535131,
0.08988041, 0.01791417, 0.02993955, 0.25796364, -0.14346326,
0.05849293, 0.12087035, -0.04232418, 0.23988574, -0.01690217,
0.13207297, -0.2849267 , 0.07666644, -0.19848674, -0.30087969,
-0.04686603])
aparc_file_lh = "/Applications/freesurfer/subjects/fsaverage/label/lh.aparc.annot"
max_thr_color = np.maximum(np.absolute(np.amin(coef_lh_parcel)),np.amax(coef_lh_parcel))
ALGO = "LinearSVC"
PARCEL = "Desikan"
surf = "white"
color = "Spectral_r"
filename = "montage.coef." + ALGO + "." + PARCEL
outdir = os.path.join("./",PARCEL,"Snapshots")
brain = Brain("fsaverage", hemi, surf, background="black", subjects_dir="/Applications/freesurfer/subjects/")
labels_lh, ctab_lh, names_lh = nib.freesurfer.read_annot(aparc_file_lh)
roi_data_lh = coef_lh_parcel
vtx_data_lh = roi_data_lh[labels_lh]
vtx_data_lh[labels_lh == -1] = -1
brain.add_data(vtx_data_lh, min=1.0e-10, mid=1.0e-09, max=max_thr_color, center=0, colormap=color, hemi="lh", transparent=True)
brain.save_montage(os.path.join(outdir,"lh." + filename + ".png"), order=['lateral', 'medial'], orientation='h', border_size=15, colorbar=[0], row=-1, col=-1)
And got the following result:
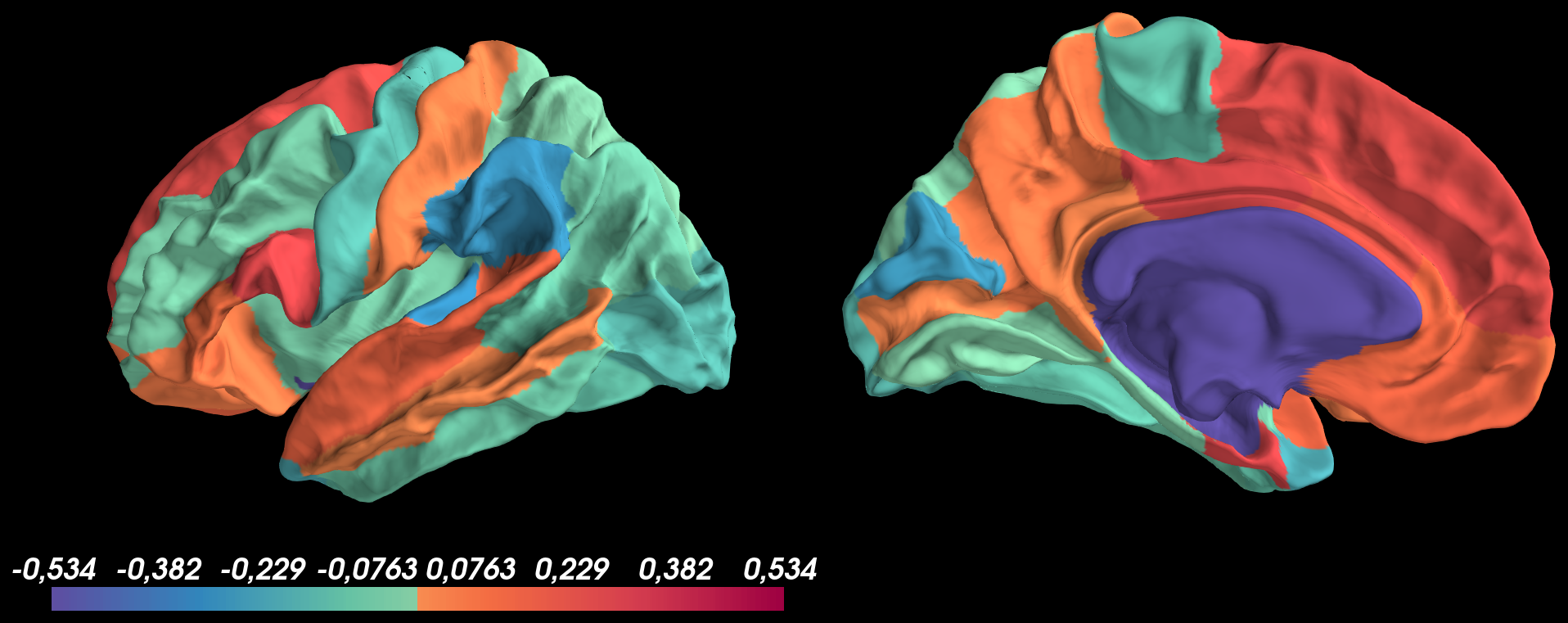
Based on this image, I don’t understand why the medial wall isn’t transparent?
Secondly, I tried to threshold both negative and positive values at thresh_low = 0.1 using pretty same code as before with the thresh option of add_data:
coef_lh_parcel = np.array([ 0.31447462, -0.08999642, 0.20214564, -0.05422508, 0. ,
-0.27036938, 0.27037478, -0.11377747, -0.07266576, -0.08349062,
0.01729273, -0.14993414, 0.04560162, -0.01732918, 0.1315399 ,
0.01141919, -0.02154149, -0.12041933, 0.26752043, 0.05535131,
0.08988041, 0.01791417, 0.02993955, 0.25796364, -0.14346326,
0.05849293, 0.12087035, -0.04232418, 0.23988574, -0.01690217,
0.13207297, -0.2849267 , 0.07666644, -0.19848674, -0.30087969,
-0.04686603])
aparc_file_lh = "/Applications/freesurfer/subjects/fsaverage/label/lh.aparc.annot"
max_thr_color = np.maximum(np.absolute(np.amin(coef_lh_parcel)),np.amax(coef_lh_parcel))
ALGO = "LinearSVC"
PARCEL = "Desikan"
surf = "white"
color = "Spectral_r"
thresh_low = 0.1
filename = "montage.coef." + ALGO + "." + PARCEL + "." + str(thresh_low)
outdir = os.path.join("/Users/matthieu/Desktop/ML_visualization",PARCEL,"Snapshots")
brain = Brain("fsaverage", hemi, surf, background="black", subjects_dir="/Applications/freesurfer/subjects/")
labels_lh, ctab_lh, names_lh = nib.freesurfer.read_annot(aparc_file_lh)
roi_data_lh = coef_lh_parcel
vtx_data_lh = roi_data_lh[labels_lh]
vtx_data_lh[labels_lh == -1] = -1
brain.add_data(vtx_data_lh, min=thresh_low, max=max_thr_color, thresh = thresh_low, center=0, colormap=color, hemi="lh")
brain.save_montage(os.path.join(outdir,"lh." + filename + ".png"), order=['lateral', 'medial'], orientation='h', border_size=15, colorbar=[0], row=-1, col=-1)
And got the following result:
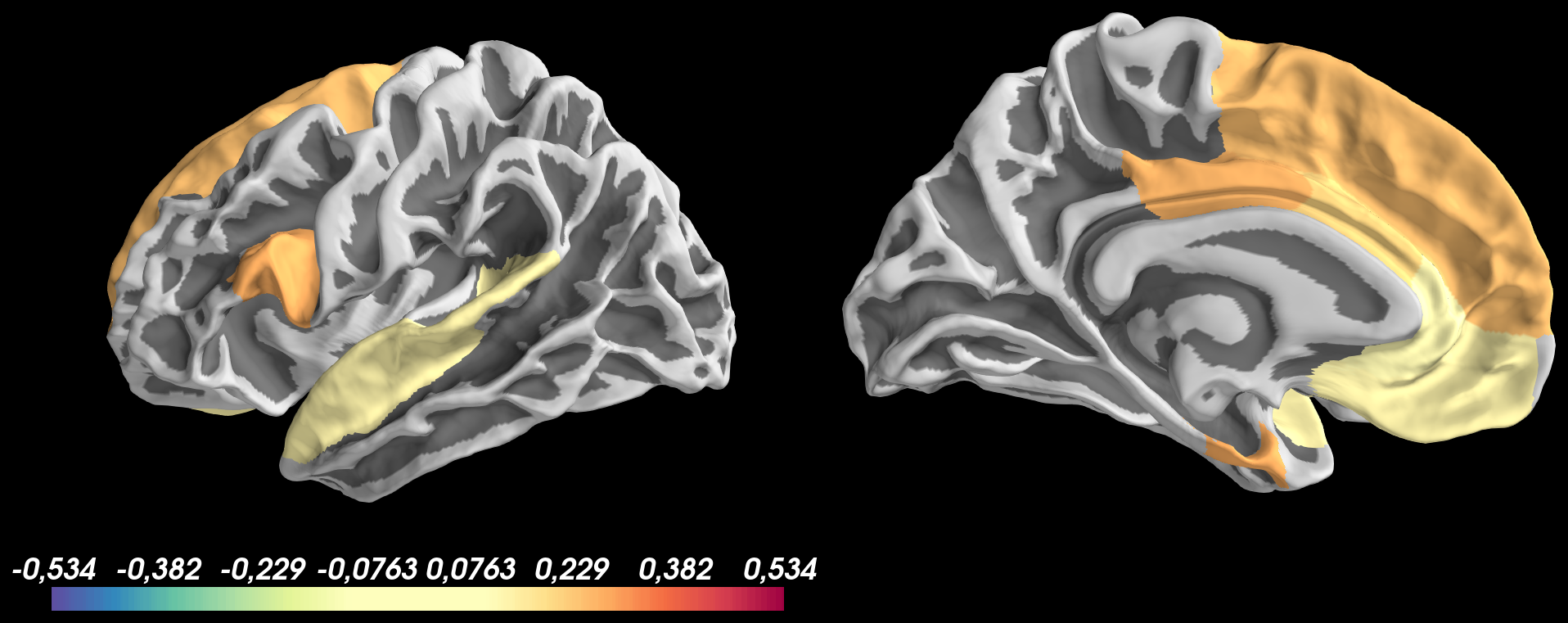
Based on this image, I don’t understand why:
- positive values are thresholded at
thresh_low = 0.1but negative values are not shown - the colormap of values > 0.1 doesn’t look good and colorscale bar isn’t updated with values from
0.1 to 0.534for positive values and-0.1 to -0.534for negative values
Issue Analytics
- State:
- Created 4 years ago
- Comments:6 (4 by maintainers)
 Top Results From Across the Web
Top Results From Across the Web
transparent medial wall and threshold both negative and positive ...
Display cortical parcellation values : transparent medial wall and threshold both negative and positive values.
Read more >Functional Parcellation of the Cerebral Cortex Across ... - NCBI
Any parcels rotated into the medial wall area when generating the null model parcellations had their homogeneity values assigned as the mean ...
Read more >1 A Multi-modal Parcellation of Human Cerebral Cortex ...
Understanding the amazingly complex human cerebral cortex requires a map. (parcellation) of its major subdivisions (cortical areas).
Read more >Issues · nipy/PySurfer
Display cortical parcellation values : transparent medial wall and threshold both negative and positive values. #290 opened on Mar 4, 2020 by mattvan83....
Read more >Hierarchical cortical gradients in somatosensory processing
Incorporating a multi-modal cortical parcellation (Glasser et al., ... Positive values indicate a contralateral preference and negative ...
Read more > Top Related Medium Post
Top Related Medium Post
No results found
 Top Related StackOverflow Question
Top Related StackOverflow Question
No results found
 Troubleshoot Live Code
Troubleshoot Live Code
Lightrun enables developers to add logs, metrics and snapshots to live code - no restarts or redeploys required.
Start Free Top Related Reddit Thread
Top Related Reddit Thread
No results found
 Top Related Hackernoon Post
Top Related Hackernoon Post
No results found
 Top Related Tweet
Top Related Tweet
No results found
 Top Related Dev.to Post
Top Related Dev.to Post
No results found
 Top Related Hashnode Post
Top Related Hashnode Post
No results found

My suggestion for your specific task is to use
threshand a specific value for the medial wall vertices but to make sure that you’re using those properly (i.e…, set the thresh below any values that appear in your data and tag the medial wall vertices with a value below that).I think the answer here is that the colormap has 256 entries and so if the distance between
centerandmidis too small, no vertex will actually get painted with that value. This behavior is surprising and I think we may want to rework the code behave closer to what is expected or, failing that, warn. cc @sbitzer, you originally added this functionality; do you have bandwidth to address this issue?Also it’s annoying that there’s no way to use
transparent=Trueto threshold out values belowminwithout getting the transparency ramp tomid, and that an exception is raised whenmin == mid.I can’t replicate this behavior on my system; the colormap maxes out at the maximum value in the data for all permutations of your examples.
Thanks for the tip about the medial wall values to 0.
I tried your advice about threshold using
center,minandtransparentas below:and got the following image:
minandmidto be transparent, but just not to display the 0 values which are in medial wall.So, I tried to add a very small mid value near min to reduce the transparent regions but keeping the transparent medial wall:
brain.add_data(vtx_data_lh, min=1.0e-10, mid=1.0e-09, max=max_thr_color, transparent=True, center=0, colormap=color, hemi="lh")But the resulting image keep displaying the medial wall even if
0<min: