`_cdf_cvm` inaccurate for small `n`
See original GitHub issueThe documentation of _cdf_cvm notes:
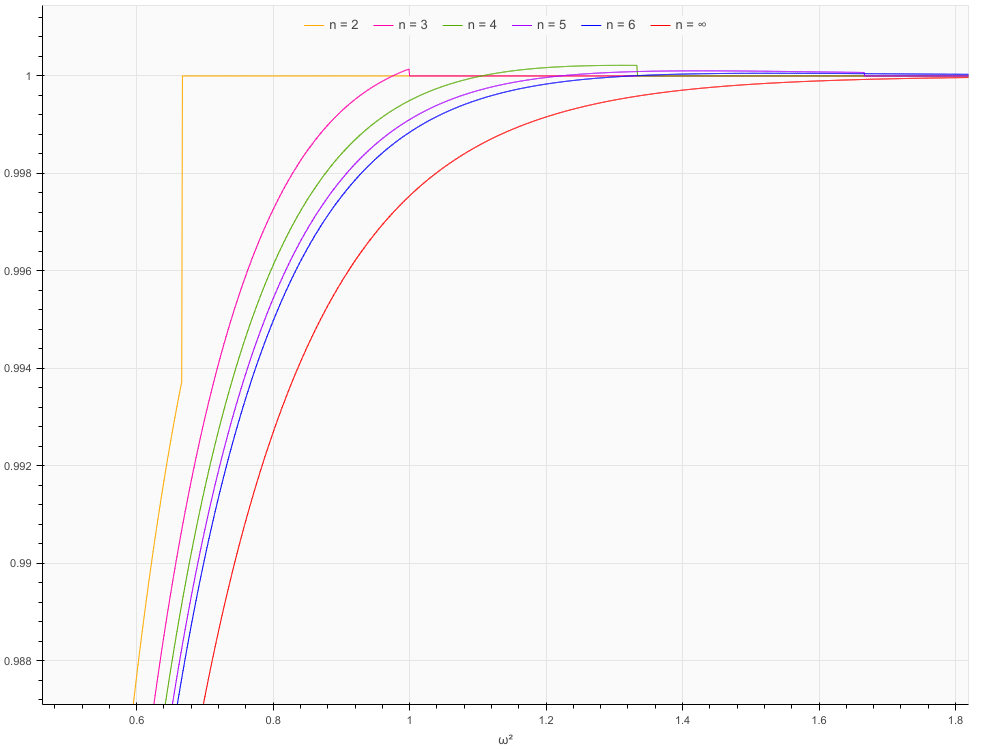
The function is not expected to be accurate for large values of x, say x > 2, when the cdf is very close to 1 and it might return values > 1 in that case, e.g. _cdf_cvm(2.0, 12) = 1.0000027556716846.
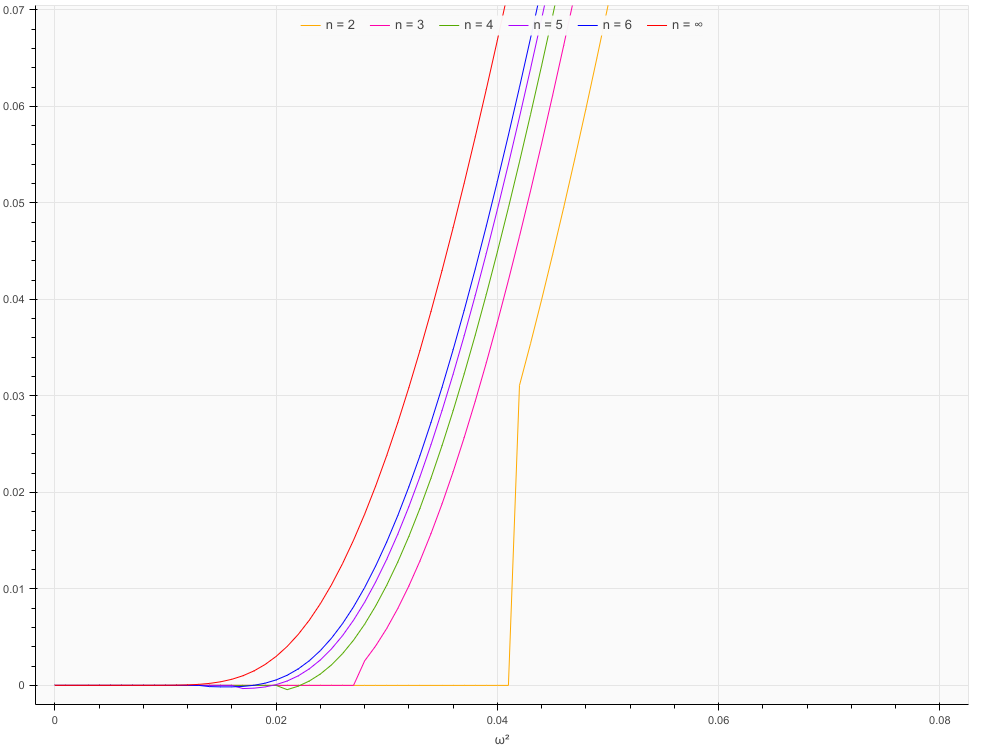
However, in addition to that, the function is inaccurate for small values of n, in particular 2. Below approx. 0.042 the value drops to 0, and above approx. 0.666 it jumps to 1. To a smaller extent, similar things happen for other small n.
I don’t know whether this can be fixed, but it should be documented.
Reproducing code example:
>>> _cdf_cvm([0.041, 0.042, 0.666, 0.667], n=2)
array([0. , 0.031081, 0.993724, 1. ])
Scipy/Numpy/Python version information:
1.6.3 1.20.2 sys.version_info(major=3, minor=8, micro=8, releaselevel='final', serial=0)
Plots
Issue Analytics
- State:
- Created 2 years ago
- Comments:10 (10 by maintainers)
 Top Results From Across the Web
Top Results From Across the Web
Problems with small sample sizes | basic statistics
In column 4, samples with n = 20 and n = 5 are very misleading. figure1. Small sample size could be less of...
Read more >Best Practices for Using Statistics on Small Sample Sizes
Some people think that if you have a small sample size you can't use statistics. Put simply, this is wrong, but it's a...
Read more >Practical Semiparametric Inference With Bayesian ...
the predictive accuracy of each candidate model hk varies across space and time, and there may exist systematic prediction bias among all hk's....
Read more >Too Much or Not Enough: Sample Sizes and Statistical Analysis
With small sample sizes, the increase in false negative rates can give a sense of complacency and lead to no process improvement.
Read more >False-positive findings and how to minimize them
In his recent paper Minimizing the changes of false positives and false negatives, Curran-Everett demonstrates that the chance of obtaining ...
Read more > Top Related Medium Post
Top Related Medium Post
No results found
 Top Related StackOverflow Question
Top Related StackOverflow Question
No results found
 Troubleshoot Live Code
Troubleshoot Live Code
Lightrun enables developers to add logs, metrics and snapshots to live code - no restarts or redeploys required.
Start Free Top Related Reddit Thread
Top Related Reddit Thread
No results found
 Top Related Hackernoon Post
Top Related Hackernoon Post
No results found
 Top Related Tweet
Top Related Tweet
No results found
 Top Related Dev.to Post
Top Related Dev.to Post
No results found
 Top Related Hashnode Post
Top Related Hashnode Post
No results found

Hi,
if you check the second reference in the documenation, you will find some more details on the approximation:
Csorgo, S. and Faraway, J. (1996). The Exact and Asymptotic Distribution of Cramér-von Mises Statistics. Journal of the Royal Statistical Society, pp. 221-234. => you can find it on the website https://people.bath.ac.uk/jjf23/papers/index.html
That is correct, the support of the exact distribution is contained in [1/(12*n), n/3], see Eq 1.1 in the article
You can take a look at Table 1 in the article, it contains a comparison of the exact values and the approximation. And indeed, for n=2, the approximation is not great but that is not a problem with the implementation but rather the way the approximation works (error is about 1/n**2, see Eq 1.8)
So if you need more accurate values for small n, there are other approaches you could use, they are briefly mentioned in the article, too. I hope it helps…
I’ve created a PR, please check.
@chrisb83:
I don’t understand the problems of the approximation well enough to say whether it would be feasible to make a more precise implementation. I suspect that would entail numerical calculations that are too expensive for the function itself, so one would probably have to pre-calculate a tabulation and then interpolate.
I agree that there is no acute problem for the GoF test, because it is unlikely that anyone would do such a test on a sample of 2 or similar, and even then at the conventional level of 0.05 one would have to Bonferroni-correct for several tests in order to reach the values where the approximation fails relevantly.
I would still appreciate if the function (either the current approximation or something better) could be exposed publicly.
scipy.specialseems like a good place for that.