Probe file not linking with spike sorters
See original GitHub issueHello,
I am using Spike Interface to run neural data through multiple sorters simultaneously. These include Kilosort2, Spyking-Circus, and Klusta
While I can read my probe file into spike interface (m15_190315_145422.prb -> given at the bottom of this message) using the load_probe_file function, the output from kilosort does not order the channels as specified by the .prb file. Also, channel 0 was intentionally excluded from the probe file geometry but it still appeared in the phy output.
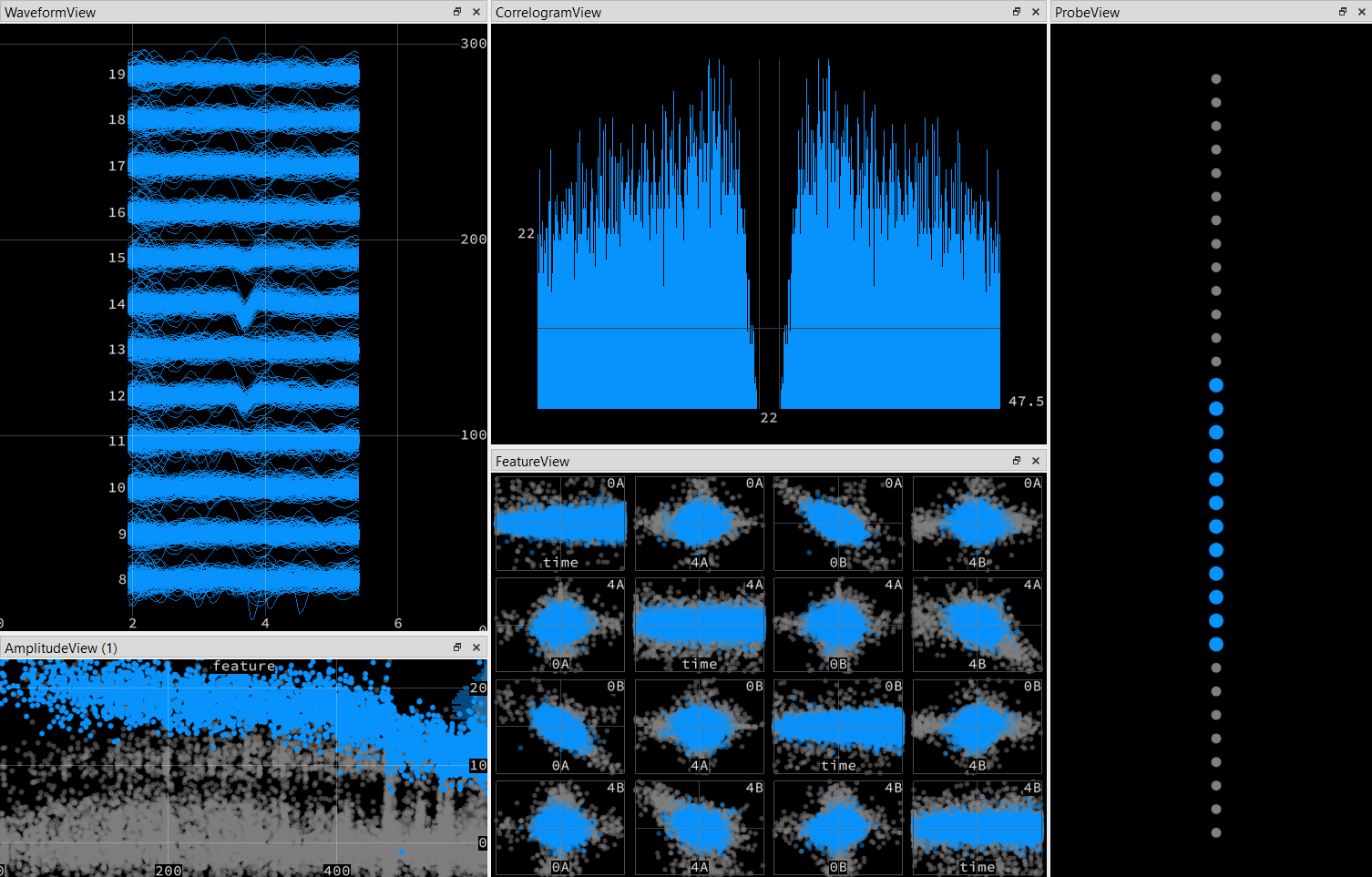
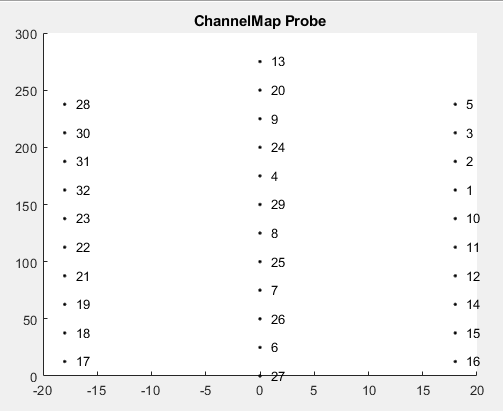
In the phy output image (image 1_PhyDisplayKs2Output), we can see selected channels for a unit template. A similar spike waveforms appear in channel 12 and 14, both of which are spatially adjacent in the probe map (see image 2_ChannelGeometry). However, channel 13 does not contain the waveform, and is also not adjacent to 12 and 14 (see image 2). Despite this, the Phy output makes me think that it using 13 still being used for this template since it’s displayed in the waveform view
my workflow is like so:
after specifying the data path in recording_folder, I then extract the data into recording
recording = se.BinDatRecordingExtractor(recording_folder + '/m15_190315_145422.dat',30000,33,'int16')
next I read in the probe file
recording.load_probe_file('m15_190315_145422.prb')
which gives the output
<spikeextractors.subrecordingextractor.SubRecordingExtractor at 0x7f02848bcb70>
I then read in my kilosort path
ss.Kilosort2Sorter.set_kilosort2_path('path')
and finally I run kilosort2 with the following:
sorting_Ks2 = ss.run_kilosort2(recording, output_folder='secondaryTest_Ks2_5422', grouping_property='group', parallel=True, verbose=True) print(f'Kilosot2 found {len(sorting_Ks2.get_unit_ids())} units')
Is this the proper way to specify the probe file or do I need to take additional steps? Thank you
I am running this through JupyterNotebook and am running an through an environment with python version 3.6.13 and running on Ubuntu version 20.04.
m15_190315_145422.prb:
total_nb_channels = 33 radius = 100
channel_groups = {
1: {‘channels’:[17,18,19,21,22,23,32,31,30,28,27,26,25,29,24,20,13,9,4,8,7,6,5,3,2,1,10,11,12,14,15,16],
‘graph’ : [],
‘geometry’: {
17: [ -18.0 , 12.65],
18: [ -18.0 , 37.65],
19: [ -18.0 , 62.65],
21: [ -18.0 , 87.65],
22: [ -18.0 , 112.65],
23: [ -18.0 , 137.65],
32: [ -18.0 , 162.65],
31: [ -18.0 , 187.65],
30: [ -18.0 , 212.65],
28: [ -18.0 , 237.65],
27: [ 0.0 , 0],
26: [ 0.0 , 50.00],
25: [ 0.0 , 100.00],
29: [ 0.0 , 150.00],
24: [ 0.0 , 200.00],
20: [ 0.0 , 250.00],
13: [ 0.0 , 275.00],
9: [ 0.0 , 225.00],
4: [ 0.0 , 175.00],
8: [ 0.0 , 125.00],
7: [ 0.0 , 75.00],
6: [ 0.0 , 25.00],
5: [ 18.0 , 237.65],
3: [ 18.0 , 212.65],
2: [ 18.0 , 187.65],
1: [ 18.0 , 162.65],
10: [ 18.0 , 137.65],
11: [ 18.0 , 112.65],
12: [ 18.0 , 87.65],
14: [ 18.0 , 62.65],
15: [ 18.0 , 37.65],
16: [ 18.0 , 12.65]}},
}
Issue Analytics
- State:
- Created 2 years ago
- Comments:21 (12 by maintainers)

 Top Related StackOverflow Question
Top Related StackOverflow Question
Great that it works! 😃
Hello, I solved the issue. It was my mistake. I repeated channel 32 twice but then I deleted channel 32 with get_slice (repeated channel and deleted channel having same index in coincidence) which somehow created the error while keeping total number of electrodes 63. Now in the fixed version total print(len(np.unique(channel_indices))) command gives 64 (so it actually gives the total number of channels before the removal of one channel I removed with get_slice).
So, it was my mistake but it was a funky mistake to debug due to the coincidences.
Thank you for all the help. All works in phy and spikeinterface_gui as well!