Unable to load .spydata files containing numpy object arrays
See original GitHub issueIssue Report Checklist
-
Searched the issues page for similar reports: https://github.com/spyder-ide/spyder/issues/10013 - same issue, but no followup
-
Read the relevant sections of the Spyder Troubleshooting Guide and followed its advice
-
Reproduced the issue after updating with
conda update spyder(orpip, if not using Anaconda) -
[X ] Could not reproduce inside
jupyter qtconsole(if console-related) (not applicable?) -
Tried basic troubleshooting (if a bug/error)
- Restarted Spyder
- Reset preferences with
spyder --reset - Reinstalled the latest version of Anaconda
- [X ] Tried the other applicable steps from the Troubleshooting Guide
-
[X ] Completed the Problem Description, Steps to Reproduce and Version sections below
Problem Description
When attempting to load .spydata files that contain numpy object arrays, i get the following error:

I believe the issue is in iofuncs.load_dictionary() https://github.com/spyder-ide/spyder/blob/9b3e311c3289adeb104d7b60c013e8b34d43b091/external-deps/spyder-kernels/spyder_kernels/utils/iofuncs.py#L415
This stack exchange post provides workaround by modifying the default for np.load() to allow_pickle=True
I’m not sure if this is legitimately a bug or, perhaps a security feature, as per this warning in the numpy documentation here

What steps reproduce the problem?
- Create a numpy array with dtype=‘object’, for example
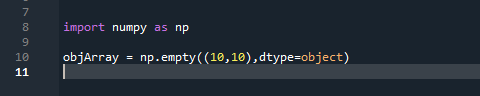
- Save environment as foo.spydata using ‘Save Data’ button above Variable Explorer Pane
- Restart spyder and attempt to load foo.spydata using ‘Import Data’ button above Variable Explorer Pane
What is the expected output? What do you see instead?
Expected out put is that the ‘objArray’ variable is imported. Instead, error above occurs “Object arrays cannot be loaded when allow_pickle=False”
Versions
- Spyder version: 4.0.1
- Python version: 3.7.3 (64-bit)
- Qt version: 5.9.6
- PyQt version: 5.9.2
- Operating System name/version: Windows 10
Dependencies
atomicwrites >=1.2.0 : 1.3.0 (OK)
chardet >=2.0.0 : 3.0.4 (OK)
cloudpickle >=0.5.0 : 1.3.0 (OK)
diff_match_patch >=20181111 : 20181111 (OK)
intervaltree : None (OK)
IPython >=4.0 : 7.12.0 (OK)
jedi =0.14.1 : 0.14.1 (OK)
nbconvert >=4.0 : 5.6.1 (OK)
numpydoc >=0.6.0 : 0.9.2 (OK)
pexpect >=4.4.0 : 4.8.0 (OK)
pickleshare >=0.4 : 0.7.5 (OK)
psutil >=0.3 : 5.6.7 (OK)
pygments >=2.0 : 2.5.2 (OK)
pylint >=0.25 : 2.4.4 (OK)
pyls >=0.31.2;<0.32.0 : 0.31.7 (OK)
zmq >=17 : 18.1.1 (OK)
qdarkstyle >=2.7 : 2.8 (OK)
qtawesome >=0.5.7 : 0.6.1 (OK)
qtconsole >=4.6.0 : 4.6.0 (OK)
qtpy >=1.5.0 : 1.9.0 (OK)
rtree >=0.8.3 : 0.9.3 (OK)
sphinx >=0.6.6 : 2.4.0 (OK)
spyder_kernels >=1.8.1;<2.0.0: 1.8.1 (OK)
watchdog : None (OK)
cython >=0.21 : 0.29.15 (OK)
matplotlib >=2.0.0 : 3.1.3 (OK)
numpy >=1.7 : 1.18.1 (OK)
pandas >=0.13.1 : 1.0.1 (OK)
scipy >=0.17.0 : 1.4.1 (OK)
sympy >=0.7.3 : 1.5.1 (OK)
Issue Analytics
- State:
- Created 4 years ago
- Comments:12 (4 by maintainers)

 Top Related StackOverflow Question
Top Related StackOverflow Question
Hi, I had this problem and the way I got around it was to convert the
numpyarray withdtype = 'object'to a list (usingnp.tolist()) and then save the environment using spyder’s interface.After using this method spyder loads the .spydata with no errors. ( I haven’t changed the
allow_pickleat all)In my case, I have used
dtype='object'to either store something other than a float ( a decimal number ) or a ragged nested sequence. so saving it as a list might be a better option anyways.This issue isn’t resolved for
Spyder version 5.3.2and1.23.3.