Strange results on 10X hgmm10k_v3 dataset
See original GitHub issueHi,
While CellBender works as expected on 10X hgmm12k (v2), on 10X hgmm10k (v3), it strangely removes large mouse gene counts and adds large human gene counts to mouse cells. 10X hgmm5k (v3) gives similar unexpected results as hgmm10k (v3). Please see logs and plots (hgmm12k and hgmm10k only) below:
hgmm12k, v2
- Log:
cellbender:remove-background: Command:
cellbender remove-background --input data/hgmm_12k/hgmm_12k_raw_gene_bc_matrices_h5.h5 --output data/cellbender/hgmm_12k_raw_gene_bc_matrices_h5.cellbender.h5 --expected-cells 12000 --total-droplets-included 22000 --epochs 150 --cuda
cellbender:remove-background: 2020-01-29 12:36:14
cellbender:remove-background: Running remove-background
cellbender:remove-background: Loading data from file data/hgmm_12k/hgmm_12k_raw_gene_bc_matrices_h5.h5
cellbender:remove-background: CellRanger v2 format
cellbender:remove-background: Trimming dataset for inference.
cellbender:remove-background: Prior on counts in empty droplets is 199
cellbender:remove-background: Prior on counts for cells is 13864
cellbender:remove-background: Excluding barcodes with counts below 159
cellbender:remove-background: Using 12000 probable cell barcodes, plus an additional 10000 barcodes, and 48062 empty droplets.
-
Elbow plot, vertical line marks --expected-cells and --total-droplets-included:
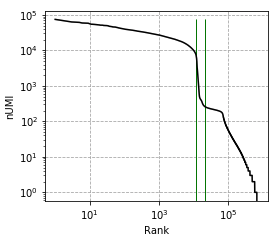
-
Before correction (called cells):
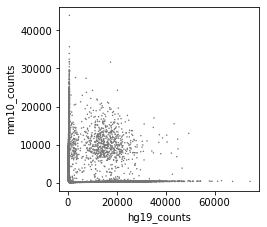
-
After correction (called cells):
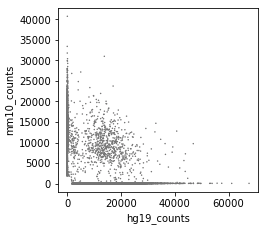
-
Convergence:
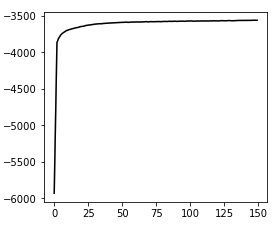
hgmm10k, v3
- Log:
cellbender:remove-background: Command:
cellbender remove-background --input data/hgmm_10k/hgmm_10k_v3_raw_feature_bc_matrix.h5 --output data/cellbender/hgmm_10k_v3_raw_feature_bc_matrix.cellbender.h5 --expected-cells 10000 --total-droplets-included 20000 --epochs 150 --cuda
cellbender:remove-background: 2020-01-29 09:31:14
cellbender:remove-background: Running remove-background
cellbender:remove-background: Loading data from file data/hgmm_10k/hgmm_10k_v3_raw_feature_bc_matrix.h5
cellbender:remove-background: CellRanger v3 format
cellbender:remove-background: Trimming dataset for inference.
cellbender:remove-background: Prior on counts in empty droplets is 444
cellbender:remove-background: Prior on counts for cells is 19036
cellbender:remove-background: Excluding barcodes with counts below 355
cellbender:remove-background: Using 10000 probable cell barcodes, plus an additional 10000 barcodes, and 56957 empty droplets.
-
Elbow plot, vertical line marks --expected-cells and --total-droplets-included:
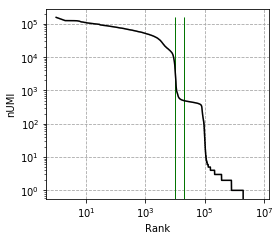
-
Before correction (called cells):
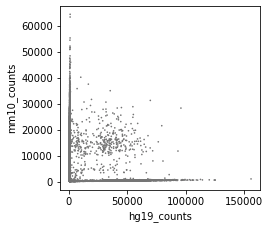
-
After correction (called cells):
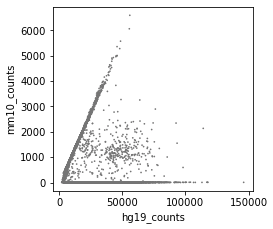
-
Convergence:
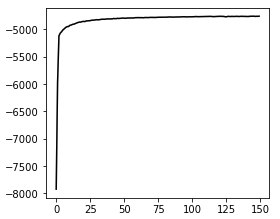
Issue Analytics
- State:
- Created 4 years ago
- Comments:9 (5 by maintainers)
 Top Results From Across the Web
Top Results From Across the Web
Datasets - 10x Genomics
Data for the Tutorial: Capturing Neutrophils in 10x Single Cell Gene Expression Data. Neutrophils are the most abundant cell type in human white...
Read more > Top Related Medium Post
Top Related Medium Post
No results found
 Top Related StackOverflow Question
Top Related StackOverflow Question
No results found
 Troubleshoot Live Code
Troubleshoot Live Code
Lightrun enables developers to add logs, metrics and snapshots to live code - no restarts or redeploys required.
Start Free Top Related Reddit Thread
Top Related Reddit Thread
No results found
 Top Related Hackernoon Post
Top Related Hackernoon Post
No results found
 Top Related Tweet
Top Related Tweet
No results found
 Top Related Dev.to Post
Top Related Dev.to Post
No results found
 Top Related Hashnode Post
Top Related Hashnode Post
No results found

This run might not have totally converged, but this is the result of running
Found it. It was coming from the use of the datatype
uint16to store gene indices during the creation of the output sparse count matrix… I guess at some point way back, I thought, “There won’t be transcriptomes with more than 65k genes, right?” Not right.I will push a fix for this soon.