scikit-misc cannot be initiated when sc.pp.highly_variable_genes(adata, n_top_genes=5000, flavor='seurat_v3')
See original GitHub issue- I have checked that this issue has not already been reported.
- I have confirmed this bug exists on the latest version of scanpy.
- (optional) I have confirmed this bug exists on the master branch of scanpy.
Hello Scanpy, When I’m running sc.pp.highly_variable_genes(adata, n_top_genes=5000, flavor=‘seurat_v3’), it asks me to install scikit-misc, which is already installed. Please see the picture below. Could you please help me to solve this issue? Thanks! Best, YJ Note: Please read this guide detailing how to provide the necessary information for us to reproduce your bug.
Minimal code sample (that we can copy&paste without having any data)
# Your code here
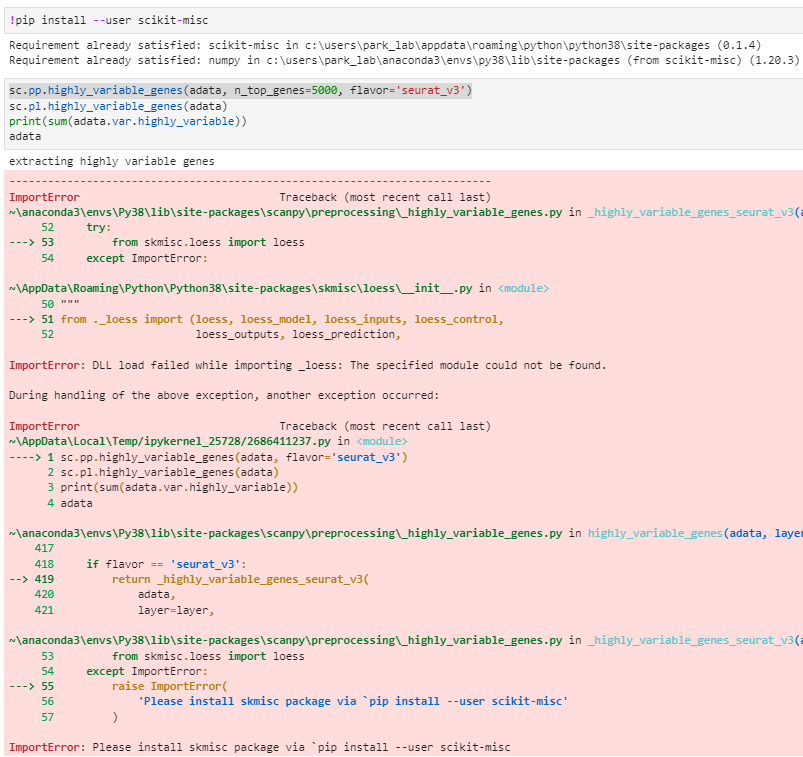
[Paste the error output produced by the above code here]
Versions
3.8
scanpy==1.8.2 anndata==0.7.8 umap==0.5.2 numpy==1.20.3 scipy==1.7.2 pandas==1.3.4 scikit-learn==1.0.1 statsmodels==0.13.1 python-igraph==0.9.8 pynndescent==0.5.5
scvelo==0.2.4 scanpy==1.8.2 anndata==0.7.8 loompy==3.0.6 numpy==1.20.3 scipy==1.7.2 matplotlib==3.5.0 sklearn==1.0.1 pandas==1.3.4
cellrank==1.5.0 scanpy==1.8.2 anndata==0.7.8 numpy==1.20.3 numba==0.54.1 scipy==1.7.2 pandas==1.3.4 pygpcca==1.0.2 scikit-learn==1.0.1 statsmodels==0.13.1 python-igraph==0.9.8 scvelo==0.2.4 pygam==0.8.0 matplotlib==3.5.0 seaborn==0.11.2
[Paste the output of scanpy.logging.print_versions() leaving a blank line after the details tag]
Issue Analytics
- State:
- Created 2 years ago
- Comments:14 (3 by maintainers)
 Top Results From Across the Web
Top Results From Across the Web
scikit-misc cannot be initiated when sc.pp ... - GitHub
Hello Scanpy, When I'm running sc.pp.highly_variable_genes(adata, n_top_genes=5000, flavor='seurat_v3'), it asks me to install scikit- ...
Read more >scanpy highly variable genes - python - Stack Overflow
The same command has no issues while working with Mac. sc.pp.highly_variable_genes(adata, layer = 'raw_data', n_top_genes = 4000, flavor = ' ...
Read more >scanpy.pp.highly_variable_genes - Read the Docs
Annotate highly variable genes [Satija15] [Zheng17] [Stuart19]. Expects logarithmized data, except when flavor='seurat_v3' , in which count data is expected.
Read more >Scikit Misc - :: Anaconda.org
Miscellaneous tools for data analysis and scientific computing. copied from cf-staging / scikit-misc · Conda · Files · Labels · Badges.
Read more > Top Related Medium Post
Top Related Medium Post
No results found
 Top Related StackOverflow Question
Top Related StackOverflow Question
No results found
 Troubleshoot Live Code
Troubleshoot Live Code
Lightrun enables developers to add logs, metrics and snapshots to live code - no restarts or redeploys required.
Start Free Top Related Reddit Thread
Top Related Reddit Thread
No results found
 Top Related Hackernoon Post
Top Related Hackernoon Post
No results found
 Top Related Tweet
Top Related Tweet
No results found
 Top Related Dev.to Post
Top Related Dev.to Post
No results found
 Top Related Hashnode Post
Top Related Hashnode Post
No results found

I found a workaround that does not require downloading the
.whlfile fornumpy=1.19.5. By default, MKL is included when you install numpy with conda. It’s good to do this in a new environment.Now I can run
sc.pp.highly_variable_genes()with no problem.Hello @davidhbrann, Thanks for the suggestion. I found that
numpy‑1.19.5+mkl‑cp36‑cp36m‑win_amd64.whlonly supports python 3.6, but the support of python 3.6 has been dropped by Scanpy. I didn’t findnumpy-1.20.3+mkl-cp38-cp38-win_amd64.whlby google yet, but I believe that your solution will work. It seems thatnumpy‑1.21.5+mkl‑cp38‑cp38‑win_amd64.whlis the only hope, but Scanpy requires numpy<=1.20. I think it’s the compatibility issue. Hope Scanpy can solve this compatibility issue in the future. Thanks! Best, YJ