ImportError: DLL load failed while importing utilsextension
See original GitHub issue- [✔] I have checked that this issue has not already been reported.
- [✔ ] I have confirmed this bug exists on the latest version of scanpy.
- [ ✔] (optional) I have confirmed this bug exists on the master branch of scanpy.
Note: Please read this guide detailing how to provide the necessary information for us to reproduce your bug.
Hello Scanpy,
I installed Scanpy, scVelo, CellRank, bbknn 2 months ago and never upgrade the packages. They were running very smoothly until I reimage my PC and reinstall Scanpy in anaconda today (Anaconda3-2021.05-Windows-x86_64, python3.8.12).
I tired pip install scanpy[leiden]. Tried conda install seaborn scikit-learn statsmodels numba pytables, conda install -c conda-forge python-igraph leidenalg. Tried installing Java and visual C++ 2012-2022 redistributable. Also tried rebuilding a new environment and reinstalled everything. Whatever I try, this bug still exists when I import Scanpy.
I guess it may be the incompatibility issue of packages. Some dependency packages which were upgraded by the developer in these months caused this incompatibility issue. Could you please help me with this bug?
Thanks!
Best,
YJ
Minimal code sample (that we can copy&paste without having any data)
import numpy as np
import pandas as pd
import scanpy as sc
import scanpy.external as sce
import scipy
sc.settings.verbosity = 3
sc.logging.print_header()
sc.set_figure_params(dpi=100, dpi_save=600)
import matplotlib.pyplot as pl
from matplotlib import rcParams
ImportError Traceback (most recent call last)
~\AppData\Local\Temp/ipykernel_7844/2696797780.py in <module>
1 import numpy as np
2 import pandas as pd
----> 3 import scanpy as sc
4 import scanpy.external as sce
5 import scipy
~\.conda\envs\Python38\lib\site-packages\scanpy\__init__.py in <module>
12 # (start with settings as several tools are using it)
13 from ._settings import settings, Verbosity
---> 14 from . import tools as tl
15 from . import preprocessing as pp
16 from . import plotting as pl
~\.conda\envs\Python38\lib\site-packages\scanpy\tools\__init__.py in <module>
15 from ._leiden import leiden
16 from ._louvain import louvain
---> 17 from ._sim import sim
18 from ._score_genes import score_genes, score_genes_cell_cycle
19 from ._dendrogram import dendrogram
~\.conda\envs\Python38\lib\site-packages\scanpy\tools\_sim.py in <module>
21 from anndata import AnnData
22
---> 23 from .. import _utils, readwrite, logging as logg
24 from .._settings import settings
25 from .._compat import Literal
~\.conda\envs\Python38\lib\site-packages\scanpy\readwrite.py in <module>
8 import pandas as pd
9 from matplotlib.image import imread
---> 10 import tables
11 import anndata
12 from anndata import (
~\.conda\envs\Python38\lib\site-packages\tables\__init__.py in <module>
43
44 # Necessary imports to get versions stored on the cython extension
---> 45 from .utilsextension import get_hdf5_version as _get_hdf5_version
46
47
ImportError: DLL load failed while importing utilsextension
Versions
Package Version
anndata 0.7.8 anyio 2.2.0 argon2-cffi 20.1.0 async-generator 1.10 attrs 21.2.0 Babel 2.9.1 backcall 0.2.0 bleach 4.1.0 Bottleneck 1.3.2 brotlipy 0.7.0 certifi 2021.10.8 cffi 1.15.0 charset-normalizer 2.0.4 colorama 0.4.4 cryptography 36.0.0 cycler 0.11.0 debugpy 1.5.1 decorator 5.1.0 defusedxml 0.7.1 entrypoints 0.3 fonttools 4.25.0 h5py 3.6.0 idna 3.3 igraph 0.9.9 importlib-metadata 4.8.2 ipykernel 6.4.1 ipython 7.29.0 ipython-genutils 0.2.0 jedi 0.18.0 Jinja2 3.0.2 joblib 1.1.0 json5 0.9.6 jsonschema 3.2.0 jupyter-client 7.1.0 jupyter-core 4.9.1 jupyter-server 1.4.1 jupyterlab 3.2.1 jupyterlab-pygments 0.1.2 jupyterlab-server 2.10.2 kiwisolver 1.3.1 leidenalg 0.8.8 llvmlite 0.37.0 MarkupSafe 2.0.1 matplotlib 3.5.0 matplotlib-inline 0.1.2 mistune 0.8.4 mkl-fft 1.3.1 mkl-random 1.2.2 mkl-service 2.4.0 mock 4.0.3 munkres 1.1.4 natsort 8.0.2 nbclassic 0.2.6 nbclient 0.5.3 nbconvert 6.1.0 nbformat 5.1.3 nest-asyncio 1.5.1 networkx 2.6.3 notebook 6.4.6 numba 0.54.1 numexpr 2.8.1 numpy 1.20.3 olefile 0.46 packaging 21.3 pandas 1.3.5 pandocfilters 1.4.3 parso 0.8.3 patsy 0.5.2 pickleshare 0.7.5 Pillow 8.4.0 pip 21.2.2 prometheus-client 0.12.0 prompt-toolkit 3.0.20 pycparser 2.21 Pygments 2.10.0 pynndescent 0.5.5 pyOpenSSL 21.0.0 pyparsing 3.0.4 pyrsistent 0.18.0 PySocks 1.7.1 python-dateutil 2.8.2 python-igraph 0.9.9 pytz 2021.3 pywin32 302 pywinpty 0.5.7 pyzmq 22.3.0 requests 2.27.1 scanpy 1.8.2 scikit-learn 1.0.2 scipy 1.7.3 seaborn 0.11.2 Send2Trash 1.8.0 setuptools 58.0.4 sinfo 0.3.4 sip 4.19.13 six 1.16.0 sniffio 1.2.0 statsmodels 0.12.2 stdlib-list 0.8.0 tables 3.6.1 terminado 0.9.4 testpath 0.5.0 texttable 1.6.4 threadpoolctl 2.2.0 tornado 6.1 tqdm 4.62.3 traitlets 5.1.1 umap-learn 0.5.2 urllib3 1.26.7 wcwidth 0.2.5 webencodings 0.5.1 wheel 0.37.1 win-inet-pton 1.1.0 wincertstore 0.2 xlrd 1.2.0 zipp 3.7.0
Issue Analytics
- State:
- Created 2 years ago
- Comments:8 (4 by maintainers)

 Top Related StackOverflow Question
Top Related StackOverflow Question
Hello @Koncopd ,
I can run the Scanpy pipeline now. However, I’m wondering whether your team did some slight changes for pca, neighboring, leiden, umap functions (not big enough to introduce a new release) inside of Scanpy v1.8.2. Because now I rerun my data and I get the UMAP like below,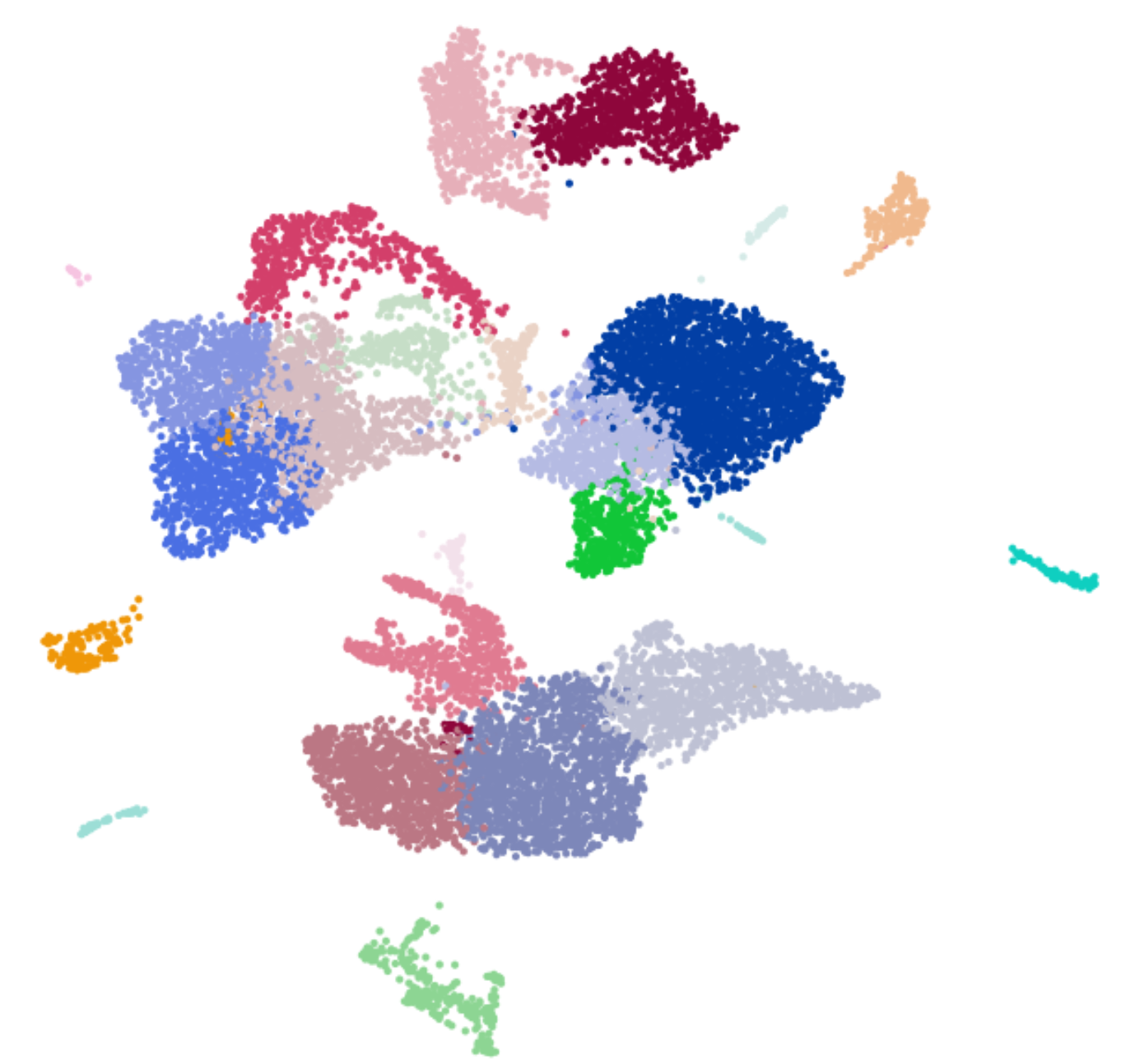
but it looks like this one month ago, run by the same version of Scanpy v1.8.2 with the same coding.
pytables 3.7.0 is incompatible with Windows. Installing pytables 3.6.1 solves the problem. For the UMAP, it’s quite weird. Whether I restart anaconda between installing individual packages plays the trick. It should be done in this way.